Astrobiologists supported in part by the NASA Astrobiology Program are applying their diverse skillset to the development of treatments for patients infected by COVID-19.
The effort centers around the work of Professor Rachel Martin at the University of California at Irvine (UC Irvine). Martin is a principal investigator (PI) with the Exobiology program and her research focuses on predicting the properties of proteases found in extremophilic microorganisms on Earth. In particular, her team studies protease in microbes from very cold environments. These microbes can help astrobiologists understand how life as we know it might be able to survive in specific locations in the outer Solar System, such as the subsurface oceans of icy moons.
“If there is liquid water on Enceladus or other places in the outer Solar System, any life that’s there would be in very cold environments,” says Martin. “We want to understand how their proteases might work.”

Doctor Rachel Martin, Professor of Chemistry and Molecular Biology & Biochemistry at UC Irvine, and Exobiology PI with the NASA Astrobiology Program.Image credit: Steve Zylius, UC Irvine.
Proteases are enzymes in cells that cut other proteins and break them down into smaller pieces. These enzymes are used in many processes, from digestion to cellular maintenance.
Proteins are made from long chains of amino acids, and proteases are able to clip those chains into smaller and smaller pieces, sometimes down to their component amino acids. For astrobiology, proteases are interesting molecules because many scientists believe that they are universal to life as we know it. As Martin puts it, “anything that has proteins has proteases,” and it is difficult to imagine life as we know it without proteins.
Martin’s team is collecting proteases from extremophiles on Earth and predicting their structural properties using molecular modeling. This allows the scientists to identify which molecules might be the most interesting to study in further detail through laboratory experiments. The research will help astrobiologists understand the minimal requirements for making a functional protease, and how these enzymes can be adapted to different environmental conditions.
Astrobiology to Virology
Earlier this year, as the global COVID-19 pandemic gained momentum, UC Irvine closed its campus along with other universities around the country. Lecture halls sat quiet as students and faculty began to shelter at home to help prevent the spread of the virus. Only activities related to COVID-19 research were allowed to continue.
As Rachel Martin finished her final in-person lecture and began preparations for closing down her laboratory, she began to wonder what her team could do to help combat the virus.
“It just seemed like a shame to not do anything when we have relevant skills,” Martin explains.
Proteases are universal to life as we know it, but these enzymes are also critical pieces in the function of a virus. In short, viral infection involves the virus entering a cell and forcing it to produce viral proteins. However, viruses are small and, compared to a cell, don’t have a lot of molecular machinery to carry out basic functions like replication. When viruses hijack cellular machinery to make proteins, they first produce one long string of amino acids known as a poly-protein. This long chain is then cut into smaller pieces using a specific viral protease, and it is these smaller pieces that make up the functional viral proteins.

A picture taken earlier this year of Marquise Crosby, a graduate student in Martin's lab at UC Irvine who is now working on coronavirus protease inhibitors.Image credit: Steve Zylius, UC Irvine.
In a healthy cell, the genes that tell the cell how to make specific proteins are written in DNA that is located in the cell’s nucleus. Those genes are transcribed into RNA, which leaves the nucleus and heads to another piece of cellular machinery called the ribosome.
“The ribosome is like a little 3-D printer for making functional proteins,” explains Martin. “It makes enzymes… it makes all of the structural proteins that our cells need.”
Coronaviruses carry RNA that mimics the cell’s RNA, and the virus takes over cellular machinery to turn its RNA into the poly-protein. Then protease moves in to chop the poly-protein into smaller pieces that make up the viral proteins. If you can stop the main protease of the virus from splicing the poly-protein into functional viral proteins, you can stop the virus dead in its tracks. Without a protease, the virus just produces one long, non-functional poly-protein, which is eventually degraded by the cell.
“It’s like a ‘PAC-MAN’ with a mouth,” explains Martin. “If we can stick something in its mouth then it can’t do its job.”
This strategy has been very successful in treating other types of viral infections, including human immunodeficiency viruses (HIV). It is important to note that HIV and SARS-CoV-2 are very different viruses, meaning that drugs like HIV protease inhibitors and Pre-Exposure Prophylaxis (PrEP) are unlikely to protect people form COVID-19. This means that specific therapeutic agents must be developed.
“When COVID-19 hit, I was thinking ‘what can we do to contribute something?” says Martin. “The thing that made the most sense was to work on the main protease of the virus.”
Martin spoke with colleagues at UC Irvine to develop a plan of action, and her lab was allowed to reopen.
“My lab was shut down for about a week,” explains Martin, “But we were one of the first labs to go back and start working on COVID-19, at least outside of the medical school.”
Martin’s lab was able to transition from research originally intended for studying extremophiles to looking at mutations in the SARS-CoV2 main protease.
“Instead of looking at different proteins from extremophiles, we’re predicting structures of variants of that main protease,” says Martin. “Thinking about astrobiology is one thing, in terms of mapping out the minimal parameters for life. In many ways, looking at a viral protease is consistent with the spirit of our original work.”

A picture taken earlier this year of Marc Sprague-Piercy, a graduate student in Martin's lab at UC Irvine who is now working on coronavirus protease inhibitors.Image credit: Steve Zylius, UC Irvine.
As with their extremophile work, the team is using molecular modeling techniques to select the most important protease targets to look at in more detail using laboratory experiments. They are able to model the structures of protease and identify areas of the molecule that could be targeted by drugs to render it inactive.
Importantly, the team is also monitoring mutations of the virus daily. So far, they are logging about two mutations a day as data comes in from around the world.
“We’re modeling every mutant of that main protease as its discovered in the clinic,” says Martin. “This helps identify end members of the spectrum of mutants.”
Drugs are screened against all the mutants and not just the original reference sequence for virus. This unique approach allows the team to look at which drugs could be broadly effective for treatments, even as the virus mutates over time.
Protease inhibitor drugs act to reduce the viral load in patients after infection, making symptoms less severe. It important to stress that protease inhibitor drugs are used after infection and are not a vaccine to protect against infection. Vaccines work to prime the immune system before exposure to a pathogen. Vaccines encourage the immune system to produce antibodies that protect a person before exposure. Protease inhibitors help to reduce the viral load after a person has been infected. Protease inhibitor drugs will be important in treating people who are infected by the virus, helping patients recover until such time as a vaccine is available.
A Large Effort
The current pandemic also means that extra measures are required in Martin’s lab to ensure the safety of staff. While the facility used to accommodate around eight researchers at a time, only two are now allowed to work in unison. A strict scheduling rotation ensures this is the case, and markings on the floor help to remind researchers in the lab to keep a safe distance from one another. Extra equipment has been borrowed from other laboratories that are currently closed so that each researcher can be designated equipment that only they come into contact with while working. The team as a whole is now only able to meet virtually over the web to discuss progress.
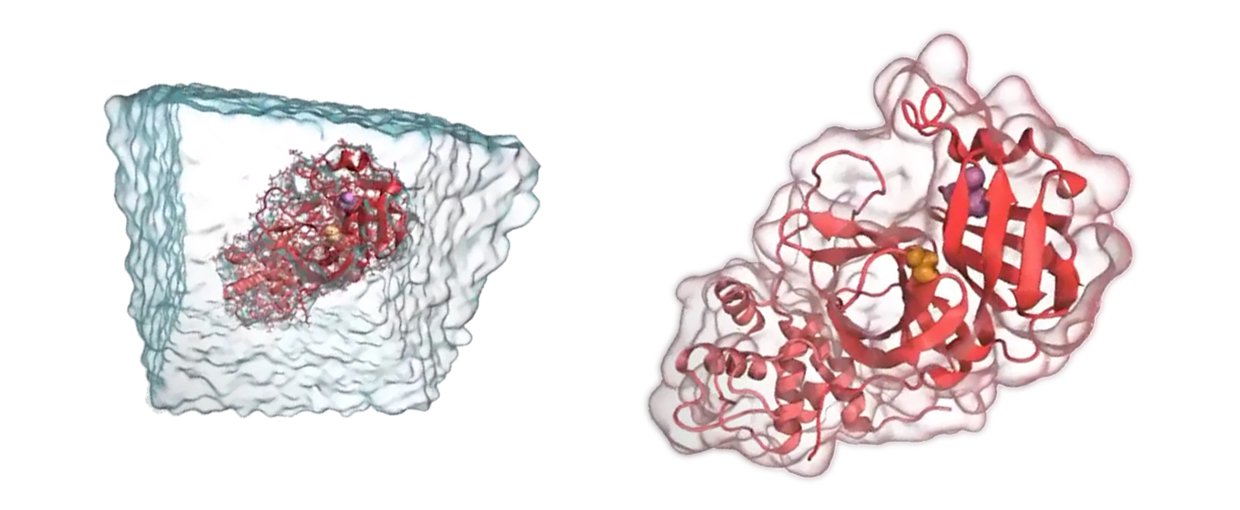
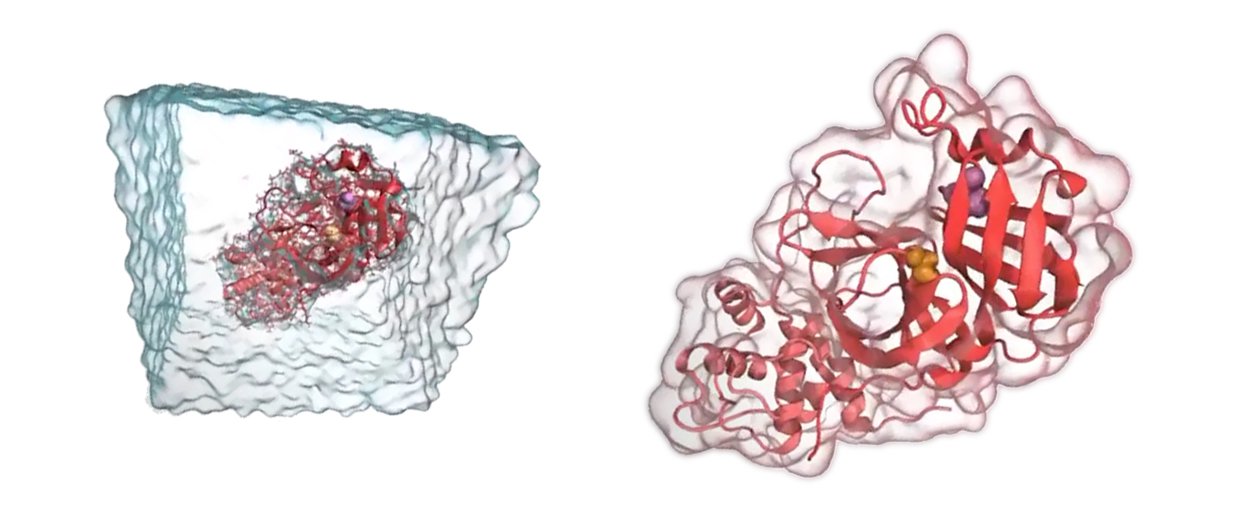
Molecular Dynamics (MD) simulations are an important tool used by the team. This technique allows researchers to simulate proteins, understand how they work, and design inhibitors before work in the lab begins.Image credit: Carter Butts, UC Irvine.
“It’s been a process getting used to it, but it’s working okay so far,” says Martin.
Martin’s lab at UC Irvine reopened in mid-March, and the project quickly gained momentum. The work now involves a large consortium of researchers, both at UC Irvine and around the world.
Professor Carter Butts of UC Irvine, also a PI on the Exobiology grant, lends expertise in computational and statistical techniques. The modeling effort is also joined by Professor Douglas Tobias in UC Irvine’s Department of Chemistry. Their work provides details about the molecules through simulations, while Martin characterizes the proteins biophysically.
The discoveries that the team makes are then used by organic chemists to actually make the drugs in the laboratory. Professor Vy Dong (UC Irvine) is leading the effort to produce the first set of protease inhibitors to test. Eight laboratories at UCI will be used for the synthetic chemistry work, and the team is now coordinating with other research groups all over the world so that molecules can be produced at a large scale without duplicating efforts.
While the protease inhibitors are being studied, Professor Jennifer Prescher (UC Irvine) and her team are performing essential tests to make sure that the inhibitors do not have any adverse effects on human cells. It is essential to make sure that an inhibitor that shuts down viral protease does not also shut down the human proteases that our cells need to function.
“This has been an absolutely unique experience for me,” says Martin. “Normally if you wanted to start a big collaboration with a whole bunch of research groups, it would take a really long time. In this case, we have put this together in about six weeks. It’s been really inspiring to me to see how scientists at UCI and all over the world have come together to work on this problem.”
Related Links:
Astrobiologists Aid in Georgia COVID-19 Test Initiative
Astrobiologists Aid in Fighting Coronavirus
NASA Response to Coronavirus (nasa.gov)
Chemistry faculty launch antiviral research project (UC Irvine)