Written byKeith Cooper

Around 4 billion years ago there lived a microbe called LUCA — the Last Universal Common Ancestor. There is evidence that it could have lived a somewhat ‘alien’ lifestyle, hidden away deep underground in iron-sulfur rich hydrothermal vents. Anaerobic and autotrophic, it didn’t breathe air and made its own food from the dark, metal-rich environment around it. Its metabolism depended upon hydrogen, carbon dioxide and nitrogen, turning them into compounds such as ammonia. Most remarkable of all, this little microbe was the beginning of a long lineage that encapsulates all life on Earth.
If we trace the tree of life far enough back in time, we come to find that we’re all related to LUCA. If the war cry for our exploration of Mars is ‘follow the water’, then in the search for LUCA it’s ‘follow the genes’. The study of the genetic tree of life, which reveals the genetic relationships and evolutionary history of organisms, is called phylogenetics. Over the last 20 years our technological ability to fully sequence genomes and build up vast genetic libraries has enabled phylogenetics to truly come of age and has taught us some profound lessons about life’s early history.
For a long time it was thought that the tree of life formed three main branches, or domains, with LUCA at the base —eukarya, bacteria and archaea. The latter two— the prokaryotes— share similarities in being unicellular and lack a nucleus, and are differentiated from one another by subtle chemical and metabolic differences. Eukarya, on the other hand, are the complex, multicellular life forms comprised of membrane-encased cells, each incorporating a nucleus containing the genetic code as well as the mitochondria ‘organelles’ powering the cell’s metabolism. The eukarya are considered so radically different from the other two branches as to necessarily occupy its own domain.

William Martin, a professor of evolutionary biology at the Heinrich Heine University in Dusseldorf, is hunting for LUCA.Image credit: Heinrich Heine University.
However, a new picture has emerged that places eukarya as an offshoot of bacteria and archaea. This “two-domain tree” was first hypothesized by evolutionary biologist Jim Lake at UCLA in 1984, but only got a foothold in the last decade, in particular due to the work of evolutionary molecular biologist Martin Embley and his lab at the University of Newcastle, UK, as well as evolutionary biologist William Martin at the Heinrich Heine University in Düsseldorf, Germany.
Bill Martin and six of his Düsseldorf colleagues (Madeline Weiss, Filipa Sousa, Natalia Mrnjavac, Sinje Neukirchen, Mayo Roettger and Shijulal Nelson-Sathi) published a 2016 paper in the journal Nature Microbiology describing this new perspective on LUCA and the two-domain tree with phylogenetics.
Ancient genes
Previous studies of LUCA looked for common, universal genes that are found in all genomes, based on the assumption that if all life has these genes, then these genes must have come from LUCA. This approach has identified about 30 genes that belonged to LUCA, but they’re not enough to tell us how or where it lived. Another tactic involves searching for genes that are present in at least one member of each of the two prokaryote domains, archaea and bacteria. This method has identified 11,000 common genes that could potentially have belonged to LUCA, but it seems far-fetched that they all did: with so many genes LUCA would have been able to do more than any modern cell can.
Bill Martin and his team realized that a phenomenon known as lateral gene transfer (LGT) was muddying the waters by being responsible for the presence of most of these 11,000 genes. LGT involves the transfer of genes between species and even across domains via a variety of processes such as the spreading of viruses or homologous recombination that can take place when a cell is placed under some kind of stress.
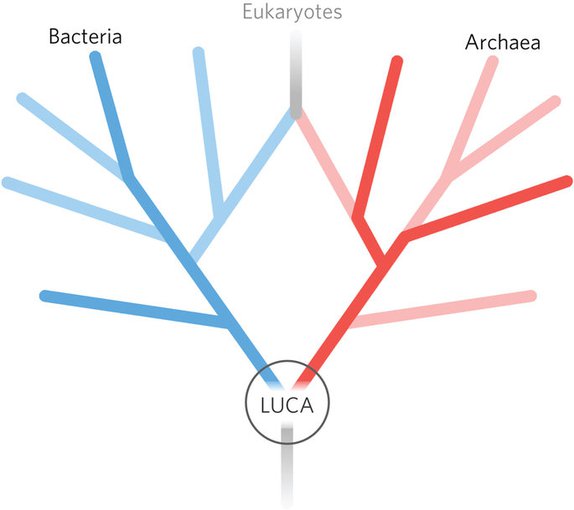
A schematic of the two-domain tree, with eukaryotes evolving from endosymbiosis between members of the two original trunks of the tree, archaea and bacteria.Image credit: Weiss et al/Nature Microbiology.
A growing bacteria or archaea can take in genes from the environment around them by ‘recombining’ new genes into their DNA strand. Often this newly-adopted DNA is closely related to the DNA already there, but sometimes the new DNA can originate from a more distant relation. Over the course of 4 billion years, genes can move around quite a bit, overwriting much of LUCA’s original genetic signal. Genes found in both archaea and bacteria could have been shared through LGT and hence would not necessarily have originated in LUCA.
Knowing this, Martin’s team searched for ‘ancient’ genes that have exceptionally long lineages but do not seem to have been shared around by LGT, on the assumption that these ancient genes should therefore come from LUCA. They laid out conditions for a gene to be considered as originating in LUCA. To make the cut, the ancient gene could not have been moved around by LGT and it had to be present in at least two groups of archaea and two groups of bacteria.
“While we were going through the data, we had goosebumps because it was all pointing in one very specific direction,” says Martin.
Once they had finished their analysis, Bill Martin’s team was left with just 355 genes from the original 11,000, and they argue that these 355 definitely belonged to LUCA and can tell us something about how LUCA lived.
Such a small number of genes, of course, would not support life as we know it, and critics immediately latched onto this apparent gene shortage, pointing out that essential components capable of nucleotide and amino acid biosynthesis, for example, were missing. “We didn’t even have a complete ribosome,” admits Martin.
However, their methodology required that they omit all genes that have undergone LTG, so had a ribosomal protein undergone LGT, it wouldn’t be included in the list of LUCA’s genes. They also speculated that LUCA could have gotten by using molecules in the environment to fill the functions of lacking genes, for example molecules that can synthesize amino acids. After all, says Martin, biochemistry at this early stage in life’s evolution was still primitive and all the theories about the origin of life and the first cells incorporate chemical synthesis from their environment.
What those 355 genes do tell us is that LUCA lived in hydrothermal vents. The Düsseldorf team’s analysis indicates that LUCA used molecular hydrogen as an energy source. Serpentinization within hydrothermal vents can produce copious amounts of molecular hydrogen. Plus, LUCA contained a gene for making an enzyme called ‘reverse gyrase’, which is found today in extremophiles existing in high-temperature environments including hydrothermal vents.

The field of hydrothermal vents known as Loki’s Castle, in the North Atlantic Ocean, where scientists found archaea believed to be related to the archaea that created eukaryotes through endosymbiosis with bacteria.Image credit: R B Pedersen/Centre for Geobiology.
Two-domain tree
Martin Embley, who specializes in the study eukaryotic evolution, says the realization of the two-domain tree over the past decade, including William Martin’s work to advance the theory, has been a “breakthrough” and has far-reaching implications on how we view the evolution of early life. “The two-domain tree of life, where the basal split is between the archaea and the bacteria, is now the best supported hypothesis,” he says.
It is widely accepted that the first archaea and bacteria were likely clostridia (anaerobes intolerant of oxygen) and methanogens, because today’s modern versions share many of the same properties as LUCA. These properties include a similar core physiology and a dependence on hydrogen, carbon dioxide, nitrogen and transition metals (the metals provide catalysis by hybridizing their unfilled electron shells with carbon and nitrogen). Yet, a major question remains: What were the first eukaryotes like and where do they fit into the tree of life?
Phylogenetics suggests that eukaryotes evolved through the process of endosymbiosis, wherein an archaeal host merged with a symbiont, in this case a bacteria belonging to the alphaproteobacteria group. In the particular symbiosis that spawned the development of eukarya, the bacteria somehow came to thrive within their archaeal host rather than be destroyed. Hence, bacteria came to not only exist within archaea but empowered their hosts to grow bigger and contain increasingly large amounts of DNA. After aeons of evolution, the symbiont bacteria evolved into what we know today as mitochondria, which are little battery-like organelles that provide energy for the vastly more complex eukaryotic cells. Consequently, eukaryotes are not one of the main branches of the tree-of-life, but merely a large offshoot.
A paper that appeared recently in Nature, written by a team led by Thijs Ettema at Uppsala University in Sweden, has shed more light on the evolution of eukaryotes. In hydrothermal vents located in the North Atlantic Ocean — centered between Greenland, Iceland and Norway, known collectively as Loki’s Castle— they found a new phylum of archaea that they fittingly named the ‘Asgard’ super-phylum after the realm of the Norse gods. The individual microbial species within the super-phylum were then named after Norse gods: Lokiarchaeota, Thorarchaeota, Odinarchaeota and Heimdallarchaeota. This super-phylum represents the closest living relatives to eukaryotes, and Ettema’s hypothesis is that eukaryotes evolved from one of these archaea, or a currently undiscovered sibling to them, around 2 billion years ago.

A hydrothermal vent in the north-east Pacific Ocean, similar to the kind of environment in which LUCA seems to have lived.Image credit: NOAA.
Closing in on LUCA
If it’s possible to date the advent of eukaryotes, and even pinpoint the species of archaea and bacteria they evolved from, can phylogenetics also date LUCA’s beginning and its split into the two domains?
It must be noted that LUCA is not the origin of life. The earliest evidence of life dates to 3.7 billion years ago in the form of stromatolites, which are layers of sediment laid down by microbes. Presumably, life may have existed even before that. Yet, LUCA’s arrival and its evolution into archaea and bacteria could have occurred at any point between 2 to 4 billion years ago.
Phylogenetics help narrow this down, but Martin Embley isn’t sure our analytical tools are yet capable of such a feat. “The problem with phylogenetics is that the tools commonly used to do phylogenetic analysis are not really sophisticated enough to deal with the complexities of molecular evolution over such vast spans of evolutionary time,” he says. Embley believes this is why the three-domain tree hypothesis lasted so long – we just didn’t have the tools required to disprove it. However, the realization of the two-domain tree suggests that better techniques are now being developed to handle these challenges.
These techniques include examining the ways biochemistry, as performed in origin-of-life experiments in the lab, can coincide with the realities of what actually happens in biology. This is a concern for Nick Lane, an evolutionary biochemist at University College of London, UK. “What I think has been missing from the equation is a biological point of view,” he says. “It seems trivially easy to make organic [compounds] but much more difficult to get them to spontaneously self-organize, so there are questions of structure that have largely been missing from the chemist’s perspective.”
For example, Lane highlights how lab experiments routinely construct the building blocks of life from chemicals like cyanide, or how ultraviolet light is utilized as an ad hoc energy source, yet no known life uses these things. Although Lane sees this as a disconnect between lab biochemistry and the realities of biology, he points out that William (Bill) Martin’s work is helping to fill the void by corresponding to real-world biology and conditions found in real-life hydrothermal vents. “That’s why Bill’s reconstruction of LUCA is so exciting, because it produces this beautiful, independent link-up with real world biology,” Lane says.
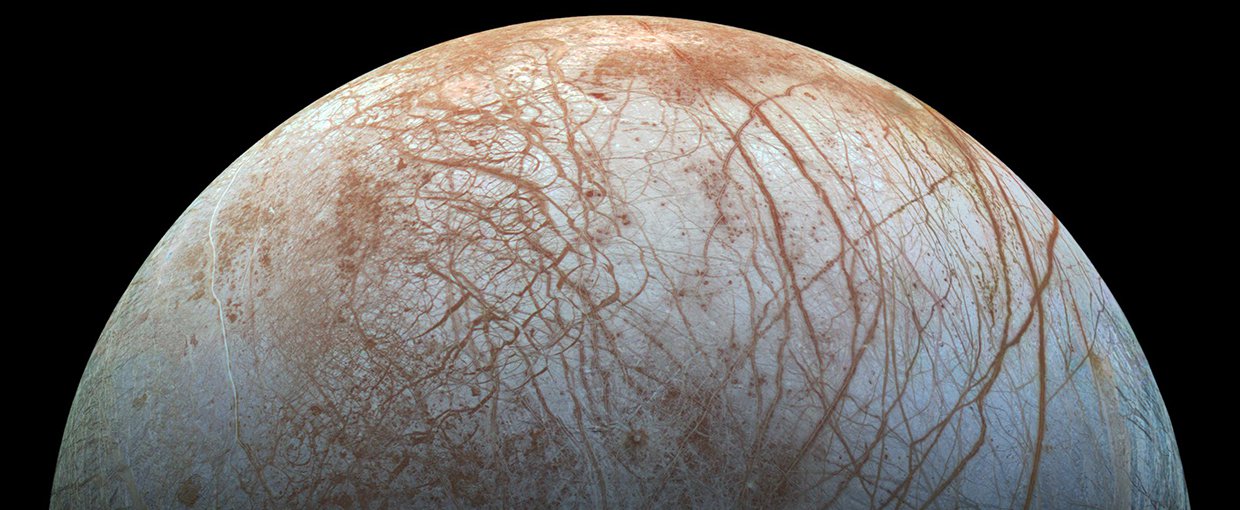
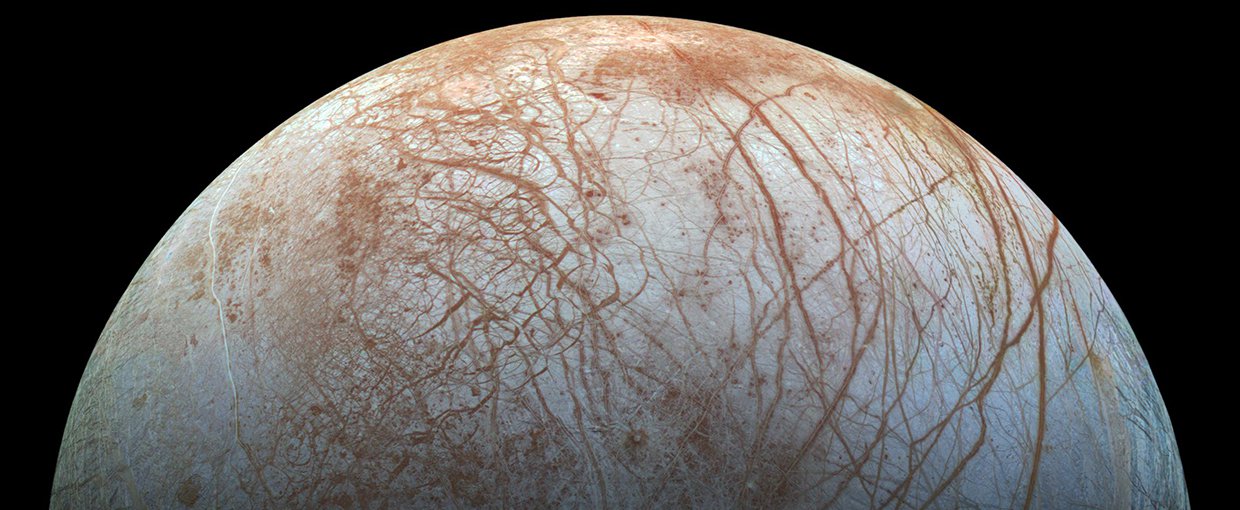
Jupiter’s moon Europa has a subterranean ocean, a rocky seabed, and geothermal heat produced by Jupiter’s gravitational tides. Water, rock and heat were all that were required by LUCA, so could similar life also exist on Europa?Image credit: NASA/JPL–Caltech/SETI Institute.
The biochemistry results in part from the geology and the materials that are available within it to build life, says Martin Embley. He sees phylogenetics as the correct tool to find the answer, citing the Wood–Ljungdahl carbon-fixing pathway as evidence for this.
Carbon-fixing involves taking non-organic carbon and turning it into organic carbon compounds that can be used by life. There are six known carbon-fixing pathways and work conducted over many decades by microbiologist Georg Fuchs at the University of Freiburg has shown that the Wood–Ljungdahl pathway is the most ancient of all the pathways and, therefore, the one most likely to have been used by LUCA. Indeed, this is corroborated by the findings of Bill Martin’s team.
In simple terms the Wood–Ljundahl pathway, which is adopted by bacteria and archaea, starts with hydrogen and carbon dioxide and sees the latter reduced to carbon monoxide and formic acid that can be used by life. “The Wood–Ljungdahl pathway points to an alkaline hydrothermal environment, which provides all the things necessary for it — structure, natural proton gradients, hydrogen and carbon dioxide,” says Martin. “It’s marrying up a geological context with a biological scenario, and it has only been recently that phylogenetics has been able to support this.”
Astrobiological implications
Understanding the origin of life and the identity of LUCA is vital not only to explaining the presence of life on Earth, but possibly that on other worlds, too. Hydrothermal vents that were home to LUCA turn out to be remarkably common within our solar system. All that’s needed is rock, water and geochemical heat. “I think that if we find life elsewhere it’s going to look, at least chemically, very much like modern life,” says Martin.
Moons with cores of rock surrounded by vast global oceans of water, topped by a thick crust of water-ice, populate the Outer Solar System. Jupiter’s moon Europa and Saturn’s moon Enceladus are perhaps the most famous, but there is evidence that hints at subterranean oceans on Saturn’s moons Titan and Rhea, as well as the dwarf planet Pluto and many other Solar System bodies. It’s not difficult to imagine hydrothermal vents on the floors of some of these underground seas, with energy coming from gravitational tidal interactions with their parent planets. The fact that the Sun does not penetrate through the ice ceiling does not matter — the kind of LUCA that Martin describes had no need for sunlight either.
“Among the astrobiological implications of our LUCA paper is the fact that you do not need light,” says Martin. “It’s chemical energy that ran the origin of life, chemical energy that ran the first cells and chemical energy that is present today on bodies like Enceladus.”
As such, the discoveries that are developing our picture of the origin of life and the existence of LUCA raise hopes that life could just as easily exist in a virtually identical environment on a distant locale such as Europa or Enceladus. Now that we know how LUCA lived, we know the signs of life to look out for during future missions to these icy moons.