2011 Annual Science Report
 Montana State University
Reporting | SEP 2010 – AUG 2011
Montana State University
Reporting | SEP 2010 – AUG 2011
Radical SAM Chemistry and Biological Ligand Accelerated Catalysis
Project Summary
A number of key reactions in biological systems are catalyzed by iron-sulfur enzymes. Iron-sulfur clusters in biology have a number of features in common with iron-sulfur minerals and their derivatives. We are using iron sulfur motifs as a model system to understand how chemistry in the abiotic mineral world was incorporated into biology on a path to the origin of life. We have found that iron-sulfur motifs in biology are synthesized and modified by reactions and mechanisms that we envision minerals could have been modified on the early prebiotic Earth. The results have had a profound impact on our ability to understand a stepwise trajectory from the nonliving to the living Earth.
Project Progress
Iron-sulfur clusters are thought to be among the most ancient cofactors in living systems. The Fe-S enzyme thrust is focused on examining the structure, mechanism, and biosynthesis of the complex Fe-S enzymes nitrogenase and hydrogenase. Exciting new results have identified important links between the biosynthesis of the H-cluster and FeMo-co and have provided direct links to the evolution of Fe-S biocatalysts from their mineral-based precursors.
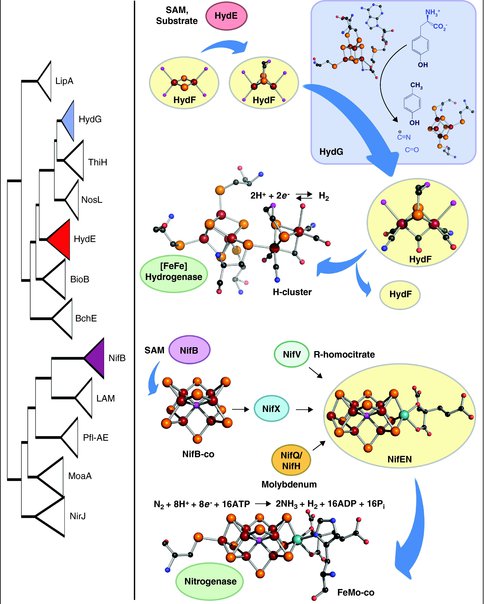
Phylogeny of radical SAM enzymes involved in complex metallocofactor assembly (left). HydE and HydG are involved in H-cluster assembly and NifB is responsible for synthesizing NifB-co, a precursor to FeMo-co. The hypothetical model for H-cluster biosynthesis in [FeFe] hydrogenase maturation (top, right). The evolutionary history of HydE and HydG, relative to other members of the radical SAM protein family, suggests that the emergence of HydE predates the emergence of HydG (left). We propose that this finding supports the hypothesis that HydE performs its chemical modifications prior to HydG during the stepwise synthesis of the H-cluster. HydE utilizes SAM and an unidentified substrate to presumably alkylate a [2Fe–2S] precursor on the scaffold HydF, followed by delivery of CO and CN− via interaction with HydG. In a final step, the 2Fe subcluster on HydF is translocated to the immature structural protein, HydAΔEFG, through the migration of the 2Fe subcluster through a cationic channel on HydA after which the movement of two loop regions close the channel. The biochemical model for FeMo-co biosynthesis in nitrogenase maturation (bottom, right). NifB utilizes SAM based radical chemistry to build NifB-co from iron–sulfur cluster precursors. NifB-co is transferred to the scaffold NifEN where molybdenum and homocitrate are introduced to synthesize FeMo-co, which is then transferred to NifDK to yield nitrogenase. Atom colors: maroon (Fe), orange (S), black©, red (O), blue (N), cyan (Mo), pink (unidentified, probably either N or O in the H-cluster and either C, N, or O in FeMo-co).
Publications
-
Broderick, J. B. (2010). Biochemistry: A radically different enzyme. Nature, 465(7300), 877–878. doi:10.1038/465877a
-
Driesener, R. C., Challand, M. R., McGlynn, S. E., Shepard, E. M., Boyd, E. S., Broderick, J. B., … Roach, P. L. (2010). -Hydrogenase Cyanide Ligands Derived From S-Adenosylmethionine-Dependent Cleavage of Tyrosine. Angewandte Chemie International Edition, 49(9), 1687–1690. doi:10.1002/anie.200907047
-
Grigoropoulos, A., & Szilagyi, R. K. (2010). Evaluation of biosynthetic pathways for the unique dithiolate ligand of the FeFe hydrogenase H-cluster. JBIC Journal of Biological Inorganic Chemistry, 15(8), 1177–1182. doi:10.1007/s00775-010-0698-y
-
Grigoropoulos, A., & Szilagyi, R. K. (2011). In silico evaluation of proposed biosynthetic pathways for the unique dithiolate ligand of the H-cluster of [FeFe]-hydrogenase. Journal of Computational Chemistry, 32(15), 3194–3206. doi:10.1002/jcc.21901
-
Harris, T. V., & Szilagyi, R. K. (2011). Comparative Assessment of the Composition and Charge State of Nitrogenase FeMo-Cofactor. Inorg. Chem., 50(11), 4811–4824. doi:10.1021/ic102446n
-
Harris, T. V., & Szilagyi, R. K. (2011). Nitrogenase Structure and Function Relationships by Density Functional Theory. Methods in Molecular Biology, None, 267–291. doi:10.1007/978-1-61779-194-9_18
-
McGlynn, S. E., Boyd, E. S., Shepard, E. M., Lange, R. K., Gerlach, R., Broderick, J. B., & Peters, J. W. (2009). Identification and Characterization of a Novel Member of the Radical AdoMet Enzyme Superfamily and Implications for the Biosynthesis of the Hmd Hydrogenase Active Site Cofactor. Journal of Bacteriology, 192(2), 595–598. doi:10.1128/jb.01125-09
-
Shepard, E. M., Boyd, E. S., Broderick, J. B., & Peters, J. W. (2011). Biosynthesis of complex iron–sulfur enzymes. Current Opinion in Chemical Biology, 15(2), 319–327. doi:10.1016/j.cbpa.2011.02.012
-
Soboh, B., Boyd, E. S., Zhao, D., Peters, J. W., & Rubio, L. M. (2010). Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Letters, 584(8), 1487–1492. doi:10.1016/j.febslet.2010.02.064
- Shepard, E.M. & Broderick, J.B. (2010). S-Adenosylmethionine and iron-sulfur clusters in biological radical reactions: The radical SAM superfamily. In: Mander, L.N. & Lui, H.W. (Eds.). Comprehensive Natural Products II: Chemistry and Biochemistry. Oxford, U.K: Elsevier Press.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
John Peters
Co-Investigator
Robert Szilagyi
Co-Investigator
Alexios Grigoropoulos
Postdoc
Eric Shepard
Postdoc
Nicholas Boswell
Doctoral Student
Ben Duffus
Doctoral Student
Shourjo Ghose
Doctoral Student
Travis Harris
Doctoral Student
Shawn McGlynn
Doctoral Student
Neelambari Joshi
Graduate Student
Alta Howells
Undergraduate Student
Amanda Byer
Unspecified Role
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.3
Origins of energy transduction