2011 Annual Science Report
 Montana State University
Reporting | SEP 2010 – AUG 2011
Montana State University
Reporting | SEP 2010 – AUG 2011
Molecular Evolution: A Top Down Approach to Examine the Origin of Key Biochemical Processes
Project Summary
The emergence of metalloenzymes capable of activating substrates such as CO, N2, and H2, were significant advancements in biochemical reactivity and in the evolution of complex life. Examples of such enzymes include [FeFe]- and [NiFe]-hydrogenase that function in H2 metabolism, Mo-, V-, and Fe-nitrogenases that function in N2 reduction, and CO dehydrogenases that function in the oxidation of CO. Many of these metalloenzymes have closely related paralogs that catalyze distinctly different chemistries, an example being nitrogenase and its closely related paralog protochlorophyllide reductase that functions in the biosynthesis of bacteriochlorophyll (photosynthesis). While the amino acid composition of these closely related paralogs are often quite similar, their biochemical reactivity and substrate specificity are often very different. This phenomenon is a direct consequence of the composition and molecular structure of the active site metallocluster, which requires a number of accessory proteins to synthesize. By specifically focusing on the origin and subsequent evolution of these metallocluster biosynthesis proteins in relation to paralogous proteins that have left clear evidence in the geological record (photosynthesis and the rise of O2), we have been able to obtain significant insight into the origin and evolution of these functional processes, and to place these events in evolutionary time.
The genomes of extant organisms provide detailed histories of key events in the evolution of complex biological processes such as CO, N2, and H2 metabolism. Advances in sequencing technology continue to increase the pace by which unique (meta)genomic data is being generated. This now makes it possible to seamlessly integrate genomic information into an evolutionary context and evaluate key events in the evolution of biological processes (e.g., gene duplications, fusions, and recruitments) within an Earth history framework. Here we describe progress in using such approaches in examining the evolution of CO, N2, and H2 metabolism.
Project Progress
The unique combination of perspectives at the ABRC that range from theoretical geochemistry and physics to biochemistry and molecular evolution has led to the development of a unique approach in molecular evolution studies. Here, the biosynthesis of active site cofactors or substrates, generated using the extensive knowledge of the biochemistry of these systems, is used to identify key events in the evolution of these pathways that lead to unique biochemical reactivities that differentiate them from closely related paralogs. These events include gene duplication, fusion, and recruitment events that can be placed within the context of knowledge of Earth history.
We have applied this approach to examine origin and evolution of nitrogenase (dinitrogen reduction), [FeFe]-hydrogenase (reversible H2 oxidation), and chlorophyll biosynthesis (proxy for photosynthesis). This endeavor has brought together complementary expertise drawn from across the NAI in target areas such as geochemistry (ASU Follow the Elements), ecology (CalTech PSARC), biochemistry (CalTech PSARC) in order to generate an integrated understanding of the evolution of nitrogenase from multiple perspectives.
ABRC scientists have also initiated several new evolutionary studies that focus on the nickel containing enzymes [NiFe]-hydrogenase (reversible H2 oxidation) and carbon monoxide dehydrogenase (CO oxidation). This study has been enhanced by collaboration with experts in metalloenyzme biochemistry from the University of Georgia, as well as experts in abiotic synthesis under hydrothermal conditions (ASU Follow the Elements). Efforts aimed at characterizing the evolutionary history of [NiFe]-hydrogenase involved in the anaerobic production of hydrogen, indicate that these genes evolved very early in the history of life.
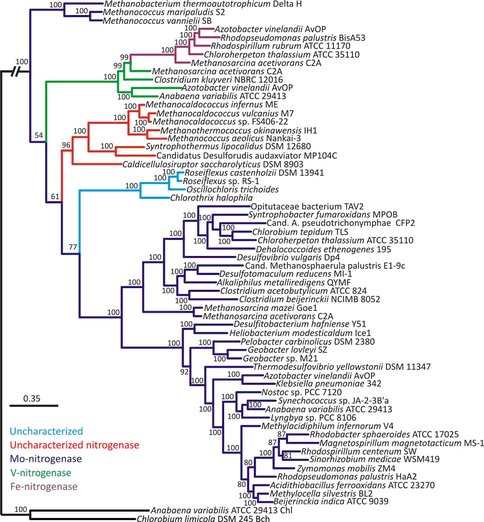
Phylogenetic tree of concatenated nitrogenase homologs (anf/nif/vnf/uncharacterized HDK. Posterior probabilities are indicated above or below nodes. Branches are colored dark blue (molybdenum-nitrogenase, Nif), green (vanadium-nitrogenase, Vnf), purple (iron-nitrogenase, Anf), red (uncharacterized nitrogenase), and light blue (uncharacterized homolog). The hash at the root was introduced to conserve space. These results strongly suggest that the oldest extant nitrogenase contained a molybdenum cofactor, with forms containing vanadium or iron evolving later.
Publications
-
Boyd, E. S., Anbar, A. D., Miller, S., Hamilton, T. L., Lavin, M., & Peters, J. W. (2011). A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology, 9(3), 221–232. doi:10.1111/j.1472-4669.2011.00278.x
-
Boyd, E. S., Hamilton, T. L., & Peters, J. W. (2011). An Alternative Path for the Evolution of Biological Nitrogen Fixation. Frontiers in Microbiology, 2. doi:10.3389/fmicb.2011.00205
-
McGlynn, S. E., Boyd, E. S., Shepard, E. M., Lange, R. K., Gerlach, R., Broderick, J. B., & Peters, J. W. (2009). Identification and Characterization of a Novel Member of the Radical AdoMet Enzyme Superfamily and Implications for the Biosynthesis of the Hmd Hydrogenase Active Site Cofactor. Journal of Bacteriology, 192(2), 595–598. doi:10.1128/jb.01125-09
-
Meuser, J. E., Boyd, E. S., Ananyev, G., Karns, D., Radakovits, R., Narayana Murthy, U. M., … Posewitz, M. C. (2011). Evolutionary significance of an algal gene encoding an [FeFe]-hydrogenase with F-domain homology and hydrogenase activity in Chlorella variabilis NC64A. Planta, 234(4), 829–843. doi:10.1007/s00425-011-1431-y
-
Mulder, D. W., Boyd, E. S., Sarma, R., Lange, R. K., Endrizzi, J. A., Broderick, J. B., & Peters, J. W. (2010). Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydAΔEFG. Nature, 465(7295), 248–251. doi:10.1038/nature08993
-
Mulder, D. W., Shepard, E. M., Meuser, J. E., Joshi, N., King, P. W., Posewitz, M. C., … Peters, J. W. (2011). Insights into [FeFe]-Hydrogenase Structure, Mechanism, and Maturation. Structure, 19(8), 1038–1052. doi:10.1016/j.str.2011.06.008
-
Shepard, E. M., Boyd, E. S., Broderick, J. B., & Peters, J. W. (2011). Biosynthesis of complex iron–sulfur enzymes. Current Opinion in Chemical Biology, 15(2), 319–327. doi:10.1016/j.cbpa.2011.02.012
-
Soboh, B., Boyd, E. S., Zhao, D., Peters, J. W., & Rubio, L. M. (2010). Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Letters, 584(8), 1487–1492. doi:10.1016/j.febslet.2010.02.064
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Eric Boyd
Co-Investigator
Joan Broderick
Co-Investigator
Shawn McGlynn
Collaborator
Eric Shepard
Postdoc
Ben Duffus
Doctoral Student
Trinity Hamilton
Doctoral Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.1
Earth's early biosphere.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms