2011 Annual Science Report
 NASA Ames Research Center
Reporting | SEP 2010 – AUG 2011
NASA Ames Research Center
Reporting | SEP 2010 – AUG 2011
Origins of Functional Proteins and the Early Evolution of Metabolism
Project Summary
The main goal of this project is to identify critical requirements for the emergence of biological complexity in early habitable environments by examining key steps in the origins and early evolution of functional proteins and metabolic reaction networks. In particular, we investigate whether protein functionality can arise from an inventory of polymers with amino acid sequences that might have naturally existed in habitable environments. We attempt the first demonstration of multiple origins of a single enzymatic function, and investigate experimentally how primordial proteins could evolve through the diversification of their structure and function. Building on this work and on our knowledge of ubiquitous proto-cellular functions and constraints of prebiotic chemistry, we conduct computer simulations aimed at elucidating fundamental principles that govern coupled evolution of early metabolic reactions and their catalysts, and transport across cell walls.
Project Progress
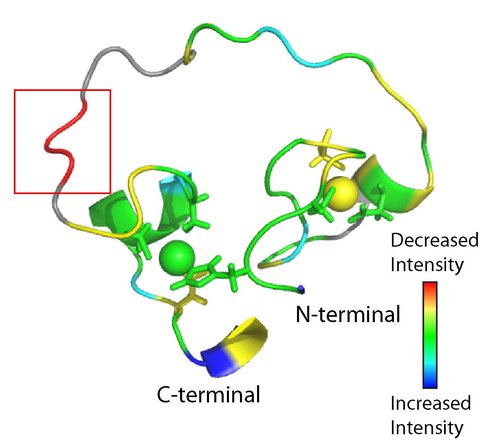
We refined the structure of the artificial RNA ligase enzyme, which was isolated from a randomized non-catalytic library of polypeptides. We showed that this enzyme exhibits a fold and catalyses a reaction not existing in nature. Even though the protein is catalytically active, its structured part is quite small and the rest of the protein consists of a flexible loop. This architecture is at variance with the basic paradigm developed for modern enzymes, which requires that functional proteins are compact. This result demonstrates that primordial enzymes, which did not undergo fine-tuning through billions of years of evolution, were different from contemporary enzymes. To probe the proposed zinc-binding sites in our enzyme (Fig. 1a), we replaced individual amino acids that could potentially coordinate zinc and tested the mutants for activity and secondary structure changes. Data on 30 mutants and titration of the enzyme with the substrate combined with the measurements of NMR loss of signal (Fig. 1b) helped to identify the potential substrate-binding site.
FIGURE 1. NMR structure of artificial RNA ligase enzyme. (a) The protein structure contains two zinc ions (displayed as balls) that are likely bound by the zinc-coordinating amino acids shown as stick models. (b) The potential substrate-binding region (red box) was identified by NMR while mixing protein and substrate. The signal intensity symbolizes the degree of change in NMR signal upon substrate binding.
We are also investigating the possibility that the same catalytic ligase activity could emerge from entirely random sequences. We have optimized the ligase selection procedure for a protein library containing 80 random amino acids and we are poised to begin the selection.
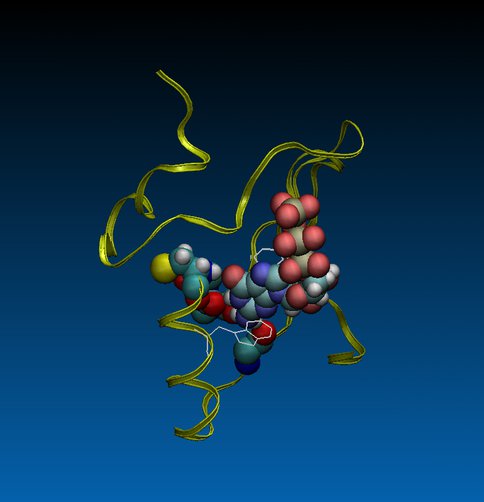
To explore how ancestral proteins evolved to acquire new functions we redesigned computationally an adenosine triphosphate (ATP)-binding protein that had evolved from random sequences of amino acids, to bind guanosine triphosphate (GTP). The redesign involved not only changing the orientation of the loop that hydrogen bonds to the nucleobase but also modifying the structures that stabilize the binding pocket and prevent water from accessing the binding site (Fig. 2)
FIGURE 2. An ATP-binding protein computationally redesigned to bind GTP, shown in ribbon and ball-and-stick representations. The two-residues (Asn and Met) binding loop in the original protein was mutated to a three-residues (Ile, Phe, and Ser) binding loop. Guanine in GTP is hydrogen bonded to the backbone nitrogen atom of cysteine 32 and oxygen atoms of isoleucine 29, phenylalanine 30, and threonine 40 (as shown in ball-and-stick representation). Side chains of tyrosine 28 and tryptophan 36 (as shown in line representation) are stacked with guanine of GTP stabilizing it in the binding pocket. Configuration is taken from a 600 ns molecular dynamics simulation on the protein in water solution.
The activity of the redesigned protein will be studied experimentally. We will also explore whether protein evolution from binding ATP to binding GTP can proceed without loss of function.
Publications
-
Seelig, B. (2011). mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nature Protocols, 6(4), 540–552. doi:10.1038/nprot.2011.312
-
Wei, C., & Pohorille, A. (2011). Permeation of Nucleosides through Lipid Bilayers. J. Phys. Chem. B, 115(13), 3681–3688. doi:10.1021/jp112104r
-
Wilson, M. A., Wei, C., Bjelkmar, P., Wallace, B. A., & Pohorille, A. (2011). Molecular Dynamics Simulation of the Antiamoebin Ion Channel: Linking Structure and Conductance. Biophysical Journal, 100(10), 2394–2402. doi:10.1016/j.bpj.2011.03.054
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Burckhard Seelig
Co-Investigator
Michael Wilson
Co-Investigator
James Lake
Collaborator
Milan Mijajlovic
Postdoc
Chenyu Wei
Research Staff
Frank Chao
Graduate Student
Aleardo Morelli
Graduate Student
Lewis Churchfield
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems