2014 Annual Science Report
 Arizona State University
Reporting | SEP 2013 – DEC 2014
Arizona State University
Reporting | SEP 2013 – DEC 2014
Task 3b: Ancient Records - Genomic
Project Summary
Task 3b team members are involved in deciphering genomic records of modern organisms as a way to understand how life on Earth evolved. At its core, this couples the integrated measurement and modeling of evolutionary mechanisms that drove the differences between extant genomes (and metagenomes), with experimental data on how environmental dynamics might have shaped these differences across geological timescales. This goal draws from team members’ expertise encompassing theoretical and computational biology, microbial evolution, and studying life in both extreme and dynamic environments across the planet.
Project Progress
In the final year of the NAI “Follow the Elements” project, team members continued their contributions to high profile and highly-cited publications. NASA/NAI support has been pivotal in advancing interdisciplinary research (detailed in the relevant sections below) between Team 3b members and NAI project collaborators, enhancing collaborations beyond the NAI initiative, and advancing the early careers of several graduate and undergraduate students. One NAI funded graduate student in Raymond’s lab (Eric Alsop) has now moved on to a Shell Oil/DOE Joint Genome Institute joint postdoctoral fellowship, a career advancement made directly possible by his research funding through the NAI “Follow the Elements” project.
Collaborators on Task 3b have been involved in several noteworthy accomplishments. Team members Boyd and Raymond contributed to an NAI graduate student first authored study by Eric Alsop integrating metagenomic and geochemical measurements around Yellowstone National Park (see figure below). This publication stood out as one of the top 10 most accessed articles for several months running in BMC Ecology. Raymond and ASU graduate student Matthew Kellom co-conducted the research for and published three papers on the isolation of several novel soil-crust inhabiting microorganisms. Chris DuPont was part of a team that finalized annotation of metal-binding protein domains in all available genomes, constructing a database of these results. This database has been leveraged in one manuscript in review, with several more in press.
Though NAI funding for these initiatives has now ceased, collaborations between team members will continue to drive new ways to integrate their interests and expertise into a stronger understanding of how the availability of key elements drove biological innovation and evolution.
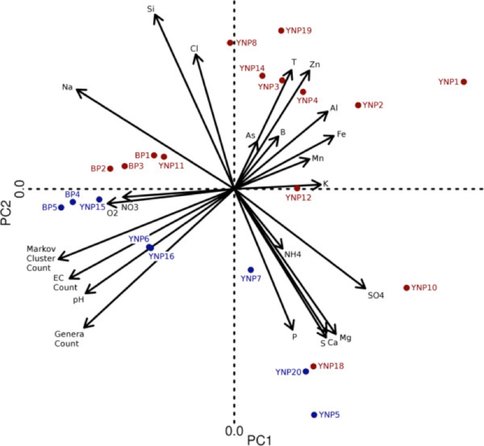
The figure below, from the NAI-supported publication by Alsop et al. (2014), shows an outstanding example of how life “follows the elements”. The biplot is generated from principal components analyses related the geochemical and diversity correlation matrices of both metagenomic and geochemical datasets, showing where hydrothermal metagenomes lie within physico-chemical space of their host hot springs. The proximal placement of biogeochemically similar sites across the PCA biplot illustrates the predictive power of PCA when applied to combined biological and geochemical sampling: predictions of biological diversity can be made directly from measurements of key (“strongly loaded”) geochemical parameters, or vice versa. Put simply, environment predicts biology.
n this figure, generated from Yellowstone National Park metagenomic and geochemical data, chemotrophic communities appear as red points and phototroph-dominated communities are shown as blue points. Elemental vectors have magnitudes and directions in line with their correlations with metagenome-derived measurements (e.g. functional and taxonomic biodiversity). Archaeal-dominated sites populate the upper right quadrant of the biplot, hinting that the geochemical parameters associated with these sites exclude bacterial life that lack functional adaptations to inhabit these geochemical extreme hot springs. Photosynthetic mat samples collected from Yellowstone National Park’s Lower Geyser Basin (BP 4, 5, YNP 6, 15 and 16) cluster at the left side of the plot, whereas photosynthetic mats found at Mammoth hot springs (YNP 5 and 20). Aquificales-dominated sites (YNP 10, 11, 12 and 13) do not cluster with each other, but instead cluster based on geographic proximity and geochemical similarity. The Bison Pool (BP) sites illustrate the strengths of PCA for correlating this multidimensional dataset: despite pronounced metagenomic differences, all five BP sites are in the same region of the biplot due to the overall similar geochemistry among sites. Further, these five sites are aligned in a linear fashion parallel to the temperature vector (due to the 32°C temperature gradient along the outflow channel of a single hot spring).
Publications
-
Alsop, E. B., Boyd, E. S., & Raymond, J. (2014). Merging metagenomics and geochemistry reveals environmental controls on biological diversity and evolution. BMC Ecology, 14(1), 16. doi:10.1186/1472-6785-14-16
-
Bailey, A. C., Kellom, M., Poret-Peterson, A. T., Noonan, K., Hartnett, H. E., & Raymond, J. (2014). Draft Genome Sequence of Bacillus sp. Strain BSC154, Isolated from Biological Soil Crust of Moab, Utah. Genome Announcements, 2(6), e01198–14–e01198–14. doi:10.1128/genomea.01198-14
-
Bailey, A. C., Kellom, M., Poret-Peterson, A. T., Noonan, K., Hartnett, H. E., & Raymond, J. (2014). Draft Genome Sequence of Massilia sp. Strain BSC265, Isolated from Biological Soil Crust of Moab, Utah. Genome Announcements, 2(6), e01199–14–e01199–14. doi:10.1128/genomea.01199-14
-
Bailey, A. C., Kellom, M., Poret-Peterson, A. T., Noonan, K., Hartnett, H. E., & Raymond, J. (2014). Draft Genome Sequence of Microvirga sp. Strain BSC39, Isolated from Biological Soil Crust of Moab, Utah. Genome Announcements, 2(6), e01197–14–e01197–14. doi:10.1128/genomea.01197-14
-
Boyd, E. S., Thomas, K. M., Dai, Y., Boyd, J. M., & Wayne Outten, F. (2014). Interplay between Oxygen and Fe–S Cluster Biogenesis: Insights from the Suf Pathway. Biochemistry, 53(37), 5834–5847. doi:10.1021/bi500488r
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Jason Raymond
Project Investigator
Christopher Dupont
Co-Investigator
Janet Siefert
Co-Investigator
Eric Alsop
Collaborator
Eric Boyd
Collaborator
Matthew Kellom
Collaborator
-
RELATED OBJECTIVES:
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments