2013 Annual Science Report
 NASA Ames Research Center
Reporting | SEP 2012 – AUG 2013
NASA Ames Research Center
Reporting | SEP 2012 – AUG 2013
Origins of Functional Proteins and the Early Evolution of Metabolism
Project Summary
The main goal of this project is to identify critical requirements for the emergence of biological complexity in early habitable environments by examining key steps in the origins and early evolution of functional proteins and metabolic reaction networks. Specifically, we investigate whether protein functionality can arise from an inventory of polypeptides that might have naturally existed in habitable environments; we attempt to demonstrate multiple origins of a single enzymatic function; we investigate how primordial proteins could evolve through the diversification of their structure and functions; and we determine how simple proteins could carry out seemingly complex functions.
Project Progress
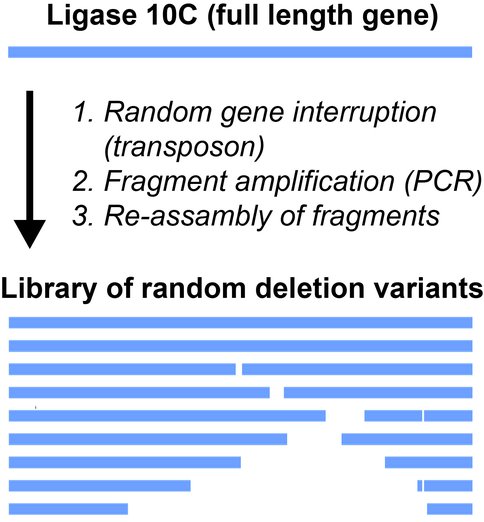
The structure and dynamics of a model primordial enzyme capable of ligating two RNA fragments was published in Nature Chemical Biology and featured on a number of science news websites. Through a combination of computer simulations and molecular biology methods we showed that mutations of several amino acids that appeared to be key for structural integrity of the enzyme did not reduce its activity, thus demonstrating a remarkable functional robustness of the ligase. We are currently searching for the most primitive version of this enzyme by selecting for activity using a ligase library with random deletions (Figure 1).
With the goal of isolating alternative RNA ligase enzymes from a library consisting of random polypeptides, we performed 13 rounds of in vitro selection but were unable to detect ligase activity. We will repeat the selection under conditions modified to include transition metal ions. These cofactors could participate in catalysis or support the three-dimensional protein structure, as we demonstrated for the previously identified ligase, which utilizes zinc.
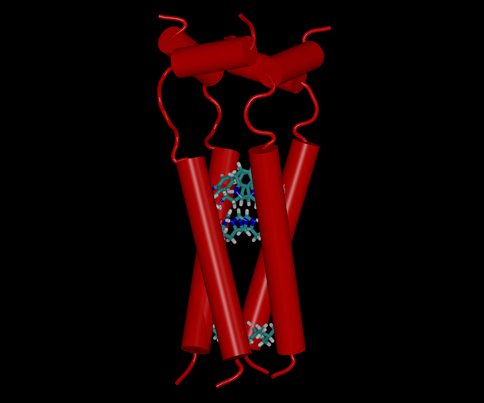
In order to understand how primordial proteins could have mediated a key cellular function – proton transport across membranous cell walls – we carried out extensive computer simulations of a simple ion channel from influenza A virus. The membrane core of the channel consists of just four identical helices (see Figure 2). Protons transported through the channel have to navigate two gates formed by valine and histidine/tryptophan amino acids, respectively; the second gate is pH-sensitive. The channel opens at this gate only when at least three of four histidines at the gate become protonated. Histidines actively participate in the transport through shuttling protons across the gate. A similar mechanism of directional proton transport might have been used in the earliest ion pumps if they contained the properly placed, photo-activated proton source.
Publications
-
Chao, F-A., Morelli, A., Iii, J. C. H., Churchfield, L., Hagmann, L. N., Shi, L., … Seelig, B. (2012). Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat Chem Biol, 9(2), 81–83. doi:10.1038/nchembio.1138
-
Golynskiy, M. V., Haugner, J. C., Morelli, A., Morrone, D., & Seelig, B. (2013). In Vitro Evolution of Enzymes. Enzyme Engineering, None, 73–92. doi:10.1007/978-1-62703-293-3_6
-
Haugner III, J. C., & Seelig, B. (2013). Universal labeling of 5′-triphosphate RNAs by artificial RNA ligase enzyme with broad substrate specificity. Chem. Commun., 49(66), 7322. doi:10.1039/c3cc44454f
-
Pohorille, A. (2012). Processes that Drove the Transition from Chemistry to Biology: Concepts and Evidence. Orig Life Evol Biosph, 42(5), 429–432. doi:10.1007/s11084-012-9304-3
-
Pohorille, A., & Pratt, L. R. (2012). Is Water the Universal Solvent for Life?. Orig Life Evol Biosph, 42(5), 405–409. doi:10.1007/s11084-012-9301-6
-
Traaseth, N. J., Chao, F-A., Masterson, L. R., Mangia, S., Garwood, M., Michaeli, S., … Veglia, G. (2012). Heteronuclear Adiabatic Relaxation Dispersion (HARD) for quantitative analysis of conformational dynamics in proteins. Journal of Magnetic Resonance, 219, 75–82. doi:10.1016/j.jmr.2012.03.024
-
Wei, C., & Pohorille, A. (2013). Activation and Proton Transport Mechanism in Influenza A M2 Channel. Biophysical Journal, 105(9), 2036–2045. doi:10.1016/j.bpj.2013.08.030
-
Wei, C., & Pohorille, A. (2013). Permeation of Aldopentoses and Nucleosides Through Fatty Acid and Phospholipid Membranes: Implications to the Origins of Life. Astrobiology, 13(2), 177–188. doi:10.1089/ast.2012.0901
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Frank Chao
Co-Investigator
Lewis Churchfield
Co-Investigator
Aleardo Morelli
Co-Investigator
Burckhard Seelig
Co-Investigator
Gareth Shannon
Co-Investigator
Chenyu Wei
Co-Investigator
Michael Wilson
Co-Investigator
James Lake
Collaborator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems