2011 Annual Science Report
 Pennsylvania State University
Reporting | SEP 2010 – AUG 2011
Pennsylvania State University
Reporting | SEP 2010 – AUG 2011
Biosignatures in Relevant Microbial Ecosystems
Project Summary
In this project, PSARC team members explore the isotope ratios, gene sequences, minerals, organic molecules, and other signatures of life in modern environments that have important similarities with early earth conditions, or with life that may be present elsewhere in the solar system and beyond. Many of these environments are “extreme” by human standards and/or have conditions that are at the limit for microbial life on Earth.
Project Progress
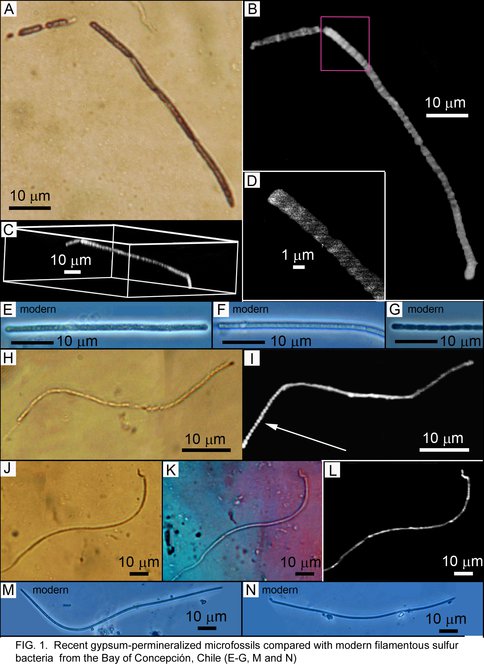
Fossil preservation in recent sulfates Schopf, Kudryavtsev, Foster (in collaboration with J. Farmer (ASU), M. Walter (UNSW))
By use of optical microscopy, confocal laser scanning microscopy, and Raman spectroscopy, we have recently discovered and documented the presence of filamentous and coccoidal fossil microorganisms permineralized in Recent gypsum deposits of Mexico, Peru, and south-central and southeastern Australia.
Recent gypsum-permineralized microfossils from Mexico compared with modern filamentous sulfur bacteria from the Bay of Concepción, Chile.
Figure 1. Gypsum-permineralization of microorganisms. Recent gypsum-permineralized microfossils from Mexico compared with modern filamentous sulfur bacteria from the Bay of Concepción, Chile.
Coupled with our discovery of diverse gypsum-permineralized microscopic fossils in Miocene (Messinian, ~6-Ma-old) deposits of northern Italy and our studies of Permian, ~255-Ma-old, sulfate deposits of Texas and New Mexico, this discovery, carried out in collaboration with J.D. Farmer (Arizona State University), has obvious relevance to the search for evidence of past life on Mars where deposits of sulfate, including gypsum, are abundant and widespread. A detailed manuscript reporting these findings, to be submitted to Astrobiology, is currently in preparation.
Molecular signatures of microbial life in the Dead Sea House, Freeman, Macalady & graduate students M. Rhodes and K. Dawson (in collaboration with A. Oren & J. R. Spear)
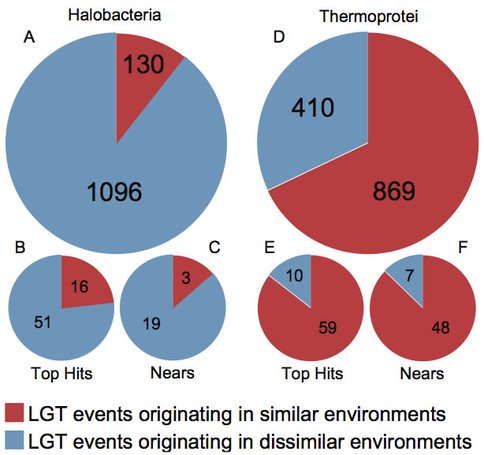
Graduate student Moshe Rhodes published a second paper for the Dead Sea project in BMC Evolutionary Biology. By investigating lateral gene transfers between Bacteria and members of the Thermoprotei and Halobacteria, we found that thermophiles appear to more commonly transfer genes with other thermophiles, while halophiles get genes from a wide range of donors. We hypothesize that hypersaline environments collect and preserve genetic material from a wide range of sources, while hot springs tend to degrade exotic DNA.
Figure 2. Origin of laterally transferred genes in the hypersaline Dead Sea.. Pie charts depicting the proportion of inter-class lateral gene transfer events from halophiles into the Halobacteria and from thermophiles into the Thermoprotei for all genes (a and d), only hits with bit scores > 500 (b and e), and only instances where multiple genes were transferred (c and f).
Figure 2. Origin of laterally transferred genes in the hypersaline Dead Sea. Pie charts depicting the proportion of inter-class lateral gene transfer events from halophiles into the Halobacteria and from thermophiles into the Thermoprotei for all genes (a and d), only hits with bit scores > 500 (b and e), and only instances where multiple genes were transferred (c and f).
A third paper recently submitted describes a series of 16S rRNA amplicon libraries constructed to span the evolution of a haloarchaeal bloom in the hypersaline Dead Sea. The analyses revealed significant population shifts among bloom populations. Furthermore, after 15 years, the 2007 inter-bloom population still retained a signature from the latter stages of the 1992 bloom.
Graduate student Katherine Dawson submitted a new paper (in revision) describing the response of archaeal halophile membrane chemistry to salinity levels. She presented strong evidence that the degree of membrane lipid unsaturation is an important adaptation to specific salinity niches in archaeal halophiles. The new findings provide a solid footing from which to unravel the contributions of archaeal membrane physiology and shifts in community composition to the organic biomarker record of salinity fluctuations in the Dead Sea (work in progress).
Peer reviewed publications:
Rhodes, M.E., Spear, J.R., Oren, A. and House, C.H., 2011. Differences in lateral gene transfer in hypersaline versus thermal environments. BMC Evolutionary Biology, 11:199, doi:10.1186/1471-2148-11-199.
Rhodes, M. E., Oren, A., and House, C. H. Revisiting Dead Sea Haloarchaeal Blooms Through High Throughput Sequencing. In review, Applied and Environmental Microbiology.
Dawson, K. S. , Freeman, K. H., and Macalady, J. L. Response of archaeal halophile membrane lipids chemistry to salinity levels. In revision, Applied & Environmental Microbiology.
Abstracts presented: Dawson, K. S.*, K. H. Freeman, and Macalady, J.L. 2011. Molecular characterization of archaeal lipids across a hypersaline gradient. Goldschmidt Conference, June 2011, Prague.
Impact cratering and subsurface microbial life Macalady, Shapiro & graduate student Korzow-Richter (in collaboration with C. Cockell, D. Vanko & M. Voytek)
Graduate student Kristine Korzow-Richter, co-advised by Beth Shapiro (primary) and Jenn Macalady, continues work on authenticated core samples from the ~2 km deep Chesapeake Bay impact structure. Because low quantities of bacteria were expected in the samples, several extraction and purification methods were tested to ensure that they were sufficient in order to detect the cells present in the samples and to remove contaminants that would interfere with downstream analyses. Five DNA extraction methods were tested. Methods tested were three standard DNA kit extraction methods: MoBio UltraClean Soil, MoBio PowerSoil, and DNA FastPrep. Additionally two phenol chloroform methods with bead beating steps were used one which was based on recent literature. Both phenol chloroform methods and the MoBio PowerSoil showed consistent positive bacterial PCR products from 104 spiked in cells. For samples with high clay content, additional steps to remove humic acids are sometimes required before PCR. We tested three purification methods: PVPP packed columns, Promega Wizard Kit, Qiagen Purification Kit. The method which showed the best removal of PCR inhibitors was the PVPP packed columns. The three best extraction methods were used on 0.5 to 5.0 g subsamples of the Eyreville core from six depths spanning from 160 to 850 meters. Each DNA extract was tested with several PCR primer sets targeting gene fragments between 100 bp and 1400 bp long. Samples with high clay content were purified with PVPP columns before PCR. None of the extracts were positive for bacterial DNA amplification. We are investigating possible causes of this negative result, including DNA extraction and amplification of marine deep sediment core samples as putative positive controls for our methods.
Methane-derived isotopic & mineralogical biosignatures in cold seeps Orphan, House, Ferry, Freeman, and McKeegan (in collaboration with J. Grotzinger)
Other current team members: Jeffrey Marlow (graduate student), Joshua Steele (postdoc), Anne Dekas (graduate student), Elizabeth Reichert (graduate student), David Case (graduate student), Grayson Chadwick (undergraduate).
Active endolithic microorganisms and methane cycling in authigenic carbonates
Methane derived authigenic carbonates have been previously shown to harbor lipid and DNA biosignatures affiliated with methanotrophic archaea and sulfate-reducing bacteria. While these biosignatures are often interpreted as ‘fossil’ remnants of microorganisms in the literature, it is also possible that they are derived from viable endolithic microorganisms. Using whole cell FISH assays group specific probes, we have recently acquired evidence of intact and active methanotrophic archaea within the interiors of authigenic carbonates from a variety of deep-sea habitats (at the seabed within active seeps and inactive areas as well as sediment hosted carbonate nodules). The aggregate morphology and abundance of these organisms varied according to habitat, with the largest archaeal biomass recorded in seafloor carbonates and nodules in proximity to active CH4 venting and approximately half as many cells in carbonates from inactive areas. In support of these observations, methane oxidation rates in these samples (assessed by the biological conversion of CH3D to D2O) followed a similar trend, with the highest rates of methanotrophy affiliated with active carbonates and nodules relative to carbonates from dormant seep areas. Stable isotope incubation experiments with 15NH4 and CH4 coupled with FISH-nanoSIMS analysis were also used to demonstrate active growth of methanotrophic consortia within the interiors of carbonate nodules. A manuscript describing the results of this work is currently in preparation (Marlow et al.).
Mineral phases associated with methanotrophic consortia.
Sulfate-dependent anaerobic oxidation of methane increases the alkalinity of the local environment, stimulating carbonate precipitation. It is currently not known whether this phenomenon is strictly mediated via the alteration of pore water alkalinity from microbial respiration or if there is a more direct role for methane-oxidizing consortia, where the microbes themselves serve as nucleation points for carbonate precipitation. Surveys of methane oxidizing microbial consortia from methane seep sediments using epifluorescence and transmitted light microscopy revealed the frequent occurrence of a translucent mineral phase associated with the exterior of many of the methanotrophic aggregates. It was hypothesized that these micro-scale crusts were comprised of calcite or aragonite, however closer examination however revealed a texture unusual for carbonates that was unaltered upon treatment with acid. Energy-dispersive X-ray spectroscopy (EDS) of individual mineral coated aggregates was employed to determine the composition of the crust and revealed the presence of C, O, Si, Al, Mg, Na, and Fe, elements suggestive of organic material in combination with aluminosilicate clay. Additional nanoSIMS 50L and electron probe analyses on prepared thin sections of these methanotrophic consortia further demonstrated the close association of clay-like mineral plates enriched in Si, Mg and Fe on the exterior of the aggregate surface.
SEM and nanoSIMS analysis of an ANME archaea and sulfate-reducing
Bacterial aggregate from methane seep sediments demonstrating
an outer shell of silicate-like material.
Figure 3. Mineral phases associated with methanotrophic consortia. SEM and nanoSIMS analysis of an ANME archaea and sulfate-reducing Bacterial aggregate from methane seep sediments demonstrating an outer shell of silicate-like material.
Future work is examining whether this phenomenon is specific to select methanotrophic archaea and sulfate-reducing bacteria and if templating of these iron-rich clays within the extracellular polymeric material is a selective or passive process and whether this exterior crust influences the preservation of these methane-oxidizing organisms and their biosignatures.
Preservation of AOM body fossils in methane-derived authigenic carbonates
With the goal of determining whether methane seep endemic microbes are preserved as microfossils in authigenic seep precipitates, we undertook a survey of Quaternary authigenic carbonates from the Eel River Basin. Petrographic examination of carbonates revealed structures that bear close morphological resemblance to the bacterial and archaeal cell aggregates thought to mediate seep carbonate precipitation in surrounding sediments. However, additional analyses using SEM, EDS, Raman spectroscopy, and rock magnetic analyses suggest that these structures are not the fossilized remains of microorganisms at all, but rather the products of the diagenetic alteration of sulfide framboids (Bailey et al., 2010). These results are important because the ability to differentiate between cellular remains and acellular mineral matter is critical for life detection efforts on other planets, as well as for tracking the evolution of biogeochemical cycles on Earth.
Anaerobic methane oxidation (AOM) coupling with sulfate and metal oxidants
Two new papers from the House lab describe AOM coupling with oxidants in the earth system. The first paper reveals that anaerobic methane oxidation rates are higher than sulfate reduction rates when sulfate concentrations are low in laboratory incubations. This suggests that the anaerobic oxidation of methane is only very weakly coupled to sulfate reduction, and that it is not always a 1 to 1 stoichiometry. The work was used to revise early Earth models of atmospheric redox during the great oxidation event based on a realization that AOM rates might have been fairly high even before sulfate became abundant in the ocean. The second paper continues our work on metal-dependent AOM, with data suggesting that the process is still based on familiar ANME taxa, and then exploring how sulfate and metal oxides might be important electron acceptors for methane oxidation on Mars.
Peer reviewed publications:
Bailey, J.V., V. Salman, G. W. Rouse, H. N. Schulz-Vogt, L. A. Levin, V. J. Orphan (2011) A dimorphic life cycle in methane seep dwelling ecotypes of the largest known bacteria. ISME J. 5:1926-1935.
Beal, E., Claire, M.W., House, C.H. 2011. The anaerobic oxidation of methane at low sulfate concentrations and the redox state of Earth’s atmosphere through time. Geobiology, 9: 131-139.
House, C.H., Beal, E.J., Orphan, V.J. 2011. The Apparent Involvement of ANMEs in Mineral Dependent Methane Oxidation, as an Analog for Possible Martian Methanotrophy. Life. 1:19-33.
Newman D, V.J. Orphan, A.L. Reysenbach (In Press) Molecular biology’s contributions to Geobiology. In Fundamentals of Geobiology. Ed. Knoll, A. D. Canfield, K. Konhauser.
Mason, O, J. Steele, T. Naehr, R. Lee, R. Thomas, J. Bailey, V. Orphan (in review) Comparison of archaeal and bacterial diversity in methane seep carbonates and host sediments, Eel River Basin and Hydrate Ridge, USA. Geobiology
Abstracts presented:
Orphan, VJ, A. Dekas, A. Green, C. House, J. Marlow, J. Steele, S. McGlynn, L. Levin (2011) Microbial partnerships and methane-oxidation in the deep sea. Goldschmidt Conference, Prague.
Marlow, J, J. Steele, B. Harrison, V. Orphan (2011) Endolithic anaerobic oxidation of methane at cold seeps. Goldschmidt Conference, Prague.
Orphan V. J. , Dekas A. E. ,Poretsky R. ,Amend J. 2010. Methane-oxidizing Archaea Fix Nitrogen in Cooperation with Sulfate-reducing Bacteria in Deep-Sea Methane Seeps. Program and Abstracts, AbSciCon10, Houston, TX.
Life in Greenland glacial ice Brenchley, Miteva, and Loveland-Curtze with undergraduate students L. Seth, N. Lazar and C. Burlingame (in collaboration with Todd Sowers)
We developed and published a new rapid and sensitive method for assessing hybridization of DNA from different organisms to estimate the relatedness of bacterial species (Loveland-Curtze J., Miteva V., Brenchley J. (2011). The new technique is based on fluorimetric detection of renaturation rates, which are used for calculating the degree of binding as an estimate of relatedness. Denatured DNA samples are reassociated under controlled conditions in a real-time PCR thermal cycler and the process is monitored fluorimetrically using SYBR Green I dye that selectively binds to double-stranded DNA. We investigated the effects of different parameters on the renaturation rates, such as the quantities of DNA and SYBR Green I used. We analyzed pairs of selected bacterial species representing different taxonomic groups and demonstrated that the method is useful for describing new species and as a screening tool to quickly identify the relatedness of uncharacterized isolates with similar 16S rRNA gene sequences.
We performed microbiological and molecular analyses of decontaminated ice core samples from the recent NEEM ice core drill in Greenland. We compared samples from different depths (100m, 644m, 1730m and 2050m) representing major ice structures types (bubbly, brittle and clathrated ice) using our established protocols for decontamination, flow cytometric cell enumeration, cultivation, DNA extraction and SSU ribosomal RNA gene phylogenetic analyses.
Cell numbers in different ice layers ranged from 104 to 106 per ml. Results showed significant diversity of the intrinsic microbial populations with variable distribution of major phylogenetic groups Proteobactera, Actinobacteria, Firmicutes, Cytophaga-Flavobacteria-Bacteroides and fungi. Parts if these results were presented at the 4th International Conference on Polar and Alpine Microbiology – Ljubljana, Slovenia and at the 2011 Penn State Undergraduate Poster Exhibition.
Peer reviewed publications:
Loveland-Curtze J., Miteva V., Brenchley J. (2011) Evaluation of a novel fluorimetric DNA-DNA hybridization method. Can. J. Microbiol., 57, 250-257.
Margesin R., Miteva V. (2011) Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 162, 346-361.
Miteva V. (2011) Microorganisms associated with glaciers. In: V. P. Singh, U. K. Haritashya and P. Singh (eds) Encyclopedia of Snow, Ice and Glaciers. Springer, pp. 741-744.
Abstracts Presented:
Miteva V., Sowers, T., Brenchley J. (2011) Authenticity of the indigenous deep ice core microbial diversity: Assessing exogenous contamination throughout the drilling process of the NEEM ice core, Greenland. 4th International Conference on Polar and Alpine Microbiology – Ljubljana, Slovenia, September 2011. (oral presentation).
Miteva V. (2011) Session Discussion Leader “Microbes in the Cryobiosphere”, Gordon Conference on Applied and Environmental Microbiology, Mount Holyoke College in South Hadley, MA, 10-14 July, 2011.
Seth L., Miteva V., Brenchley J. (2011) Diversity among small-subunit rRNA gene clones obtained from Estisol 240 and Coasol based drilling fluid and post coring chips from a Greenland glacier ice core. Penn State Undergraduate Poster Exhibition, 12-13 April, 2011.
Outreach activity:
Teacher’s Workshop (2011) Brenchley J., Loveland-Curtze J., Miteva V., L. Liermann and student N. Lazar organized laboratory activity related to the topic: Life on cold planets (Europa and Enceladus) and Earth glacial ice.
Biosignatures of life in Archean-analog anoxic biofilms Macalady & graduate students McCauley, Jones (in collaboration with A. Montanari, A. Zerkle, J. Farquhar)
Astrobiology Dual Title graduate student Rebecca McCauley is characterizing microbial life in anoxic cave lakes ~600 m below ground surface using genetic and geochemical approaches. Deep sequencing of 16S rRNA amplicons from the environment (“pyrotags”) confirm earlier hypotheses that archaea are of minor importance, and that sulfate reduction is one of the primary metabolisms. Genes from multiple pathways for autotrophic metabolism (carbon fixation), including RuBisCO, are present as well as diverse copies of dsrAB, the central gene in sulfate reduction. Large metagenomic datasets for several biofilm samples collected in this environment are nearing completion at DOE Joint Genome Institute, and are expected to shed new light on the metabolic attributes of the major populations. The major and minor sulfur isotope systematics of the cave ecosystem are being investigated by Zerkle (Univ. of Newcastle) and Farquhar (Univ. of Maryland).
Abstracts presented:
McCauley, R. L.*, Jones, D.S., Schaperdoth, I. and Macalady, J. L.. 2011. Lago Infinito (Frasassi Caves, Italy) Microbial Community as an Analog for pre-Phototrophic Earth. Astrobiology Graduate Conference, May 2011. Astrobiology 11(4): 367-389. doi:10.1089/ast.2011.1400.
Zerkle, A. L.*, Macalady, J.L., Jones, D.S., and Farquhar, J. 2011. S isotope investigation of a redox-stratified system dominated by chemotrophic sulfide oxidation. Goldschmidt Meeting, Prague.
McCauley, R. L.*, Jones, D. S., Schaperdoth, I., Steinberg, L. and Macalady, J. L.. 2010. Metabolic strategies in energy-limited microbial communities in the anoxic subsurface (Frasassi cave system, Italy). Abstract B21B-0317 presented at 2010 Fall Meeting, AGU, San Francisco, Calif., 13-17 Dec. Eos Transactions. AGU Fall Meeting Supplement.
Biosignatures of life in ancient stratified ocean analogs Macalady, Freeman, & Kump with graduate students J. Fulton, K. Meyer & R. McCauley (in collaboration with P. Welander (MIT), R. Summons (MIT), D. Newman (Caltech))
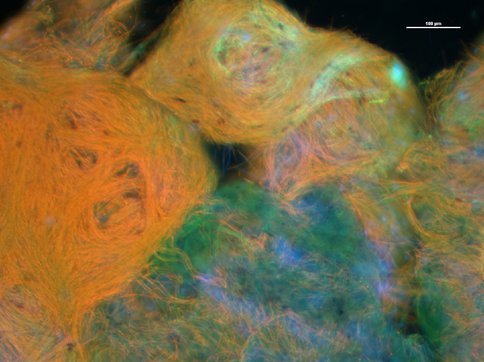
Instigated by Macalady, Kump and National Geographic Explorer Kenneth Broad (University of Miami) in 2010, this project investigates biosignatures of life in modern analogs for stratified ancient and/or extraterrestrial oceans. Seed funding provided in 2010 by National Geographic/NOVA resulted in a preliminary dataset for stratified sinkholes in the Bahamas (see publication) and NOVA/National Geographic television and magazine coverage. A field workshop at stratified Fayetteville Green Lake sponsored by NAI, Agouron Institute, and CIFAR was attended by 50 national and international participants. Emerging from the workshop, a special issue of Geobiology devoted to biosignatures in anaerobic photic ecosystems is nearing completion, edited by Macalady and colleagues Julia Maresca (U. Delaware) and Sean Crowe (U. of Southern Denmark). A new collaboration between the PSARC team and the MIT team was initiated to study the genetics and environmental controls on hopanoid production (abstract below).
Autofluorescence from photosynthetic pigments in a sinkhole biofilm that makes abundant biomarker hopanoids. The most abundant populations in the biofilm are cyanobacteria and green sulfur bacteria.
Figure 4. Hopanoid-producing phototrophs. Autofluorescence from photosynthetic pigments in a sinkhole biofilm that makes abundant biomarker hopanoids. The most abundant populations in the biofilm are cyanobacteria and green sulfur bacteria.
A website monitoring the activities of an informal working group on Early Earth Photosynthesis is maintained by Macalady (see http://www.geosc.psu.edu/~jlm80/EEP.html).
Peer reviewed publications:
Gonzalez, B. C., Iliffe, T. M., Macalady, J. L., Schaperdoth, I., and Kakuk, B. Microbial hotspots in anchialine blue holes: initial discoveries from the Bahamas. Hydrobiologia 677:149-156.
Meyer, K. , Freeman, K. H. , Macalady, J. L. , Fulton, J., Schaperdoth, I., and Kump, L. 2011. Carotenoid biomarkers as an imperfect reflection of the anaerobic phototrophic community in meromictic Fayetteville Green Lake. Geobiology 9:321-329.
Abstracts presented:
Macalady, J.L.* [invited], Albrecht H, Welander P, Fulton J, Schaperdoth I, White T, Freeman K, Newman D & Summons R. 2011. Proterozoic analog ecosystem and organic biomarkers in a Florida sinkhole. Goldschmidt Meeting, Prague.
Publications
-
Bailey, J. V., Salman, V., Rouse, G. W., Schulz-Vogt, H. N., Levin, L. A., & Orphan, V. J. (2011). Dimorphism in methane seep-dwelling ecotypes of the largest known bacteria. ISME J, 5(12), 1926–1935. doi:10.1038/ismej.2011.66
-
Beal, E. J., Claire, M. W., & House, C. H. (2011). High rates of anaerobic methanotrophy at low sulfate concentrations with implications for past and present methane levels. Geobiology, None, no–no. doi:10.1111/j.1472-4669.2010.00267.x
-
Gonzalez, B. C., Iliffe, T. M., MacAlady, J. L., Schaperdoth, I., & Kakuk, B. (2011). Microbial hotspots in anchialine blue holes: initial discoveries from the Bahamas. Hydrobiologia, 677(1), 149–156. doi:10.1007/s10750-011-0932-9
-
House, C. H., Beal, E. J., & Orphan, V. J. (2011). The Apparent Involvement of ANMEs in Mineral Dependent Methane Oxidation, as an Analog for Possible Martian Methanotrophy. Life, 1(1), 19–33. doi:10.3390/life1010019
-
Loveland-Curtze, J., Miteva, V. I., & Brenchley, J. E. (2011). Evaluation of a new fluorimetric DNA–DNA hybridization method. Canadian Journal of Microbiology, 57(3), 250–255. doi:10.1139/w10-121
-
Margesin, R., & Miteva, V. (2011). Diversity and ecology of psychrophilic microorganisms. Research in Microbiology, 162(3), 346–361. doi:10.1016/j.resmic.2010.12.004
-
Meyer, K. M., MacAlady, J. L., Fulton, J. M., Kump, L. R., Schaperdoth, I., & Freeman, K. H. (2011). Carotenoid biomarkers as an imperfect reflection of the anoxygenic phototrophic community in meromictic Fayetteville Green Lake. Geobiology, 9(4), 321–329. doi:10.1111/j.1472-4669.2011.00285.x
-
Newman, D. K., Orphan, V. J., & Reysenbach, A-L. (2012). Molecular Biology’s Contributions to Geobiology. Fundamentals of Geobiology, None, 228–249. doi:10.1002/9781118280874.ch13
-
Rhodes, M. E., Spear, J. R., Oren, A., & House, C. H. (2011). Differences in lateral gene transfer in hypersaline versus thermal environments. BMC Evolutionary Biology, 11(1), 199. doi:10.1186/1471-2148-11-199
- Dawson, K.S., Freeman, K.H. & MacAlady, J.L. (2011). Molecular characterization of archaeal lipids across a hypersaline gradient. Goldschmidt Conference. Prague.
- Dawson, K.S., Freeman, K.H. & MacAlady, J.L. (In Revision). Response of archaeal halophile membrane lipids chemistry to salinity levels. Applied & Environmental Microbiology.
- MacAlady, J.L., Albrecht, H., Welander, P., Fulton, J., Schaperdoth, I., White, T., Freeman, K., Newman, D. & Summons, R. (2011). Proterozoic analog ecosystem and organic biomarkers in a Florida sinkhole. Goldschmidt Meeting. Prague.
- Marlow, J., Steele, J., Harrison, B. & Orphan, V. (2011). Endolithic anaerobic oxidation of methane at cold seeps. Goldschmidt Conference. Prague.
- Mason, O, J.S., Naehr, T., Lee, R., Thomas, R., Bailey, J. & Orphan, V. (In Review). Comparison of archaeal and bacterial diversity in methane seep carbonates and host sediments, Eel River Basin and Hydrate Ridge, USA. Geobiology.
- McCauley, R.L., Jones, D.S., Schaperdoth, I. & MacAlady, J.L. (2011). Lago Infinito (Frasassi Caves, Italy) Microbial Community as an Analog for pre-Phototrophic Earth. Astrobiology, 11(4): 367-389. doi:10.1089/ast.2011.1400
- McCauley, R.L., Jones, D.S., Schaperdoth, I., Steinberg, L. & MacAlady, J.L. (2010). Metabolic strategies in energy-limited microbial communities in the anoxic subsurface. AGU Fall Meeting Supplement. San Francisco, CA.
- Miteva, V. (2011). Microorganisms associated with glaciers. In: V. P. Singh, U.K.H.a.P.S. (Eds.). Encyclopedia of Snow, Ice and Glaciers. Springer.
- Miteva, V. (2011). Session Discussion Leader “Microbes in the Cryobiosphere”. Gordon Conference on Applied and Environmental Microbiology. Mount Holyoke College in South Hadley, MA.
- Miteva, V., Sowers, T. & Brenchley, J. (2011). Authenticity of the indigenous deep ice core microbial diversity: Assessing exogenous contamination throughout the drilling process of the NEEM ice core. Greenland. 4th International Conference on Polar and Alpine Microbiology. Ljubljana, Slovenia.
- Orphan, V., Dekas, A., Green, A., House, C., Marlow, J., Steele, J., McGlynn, S. & Levin, L. (2011). Microbial partnerships and methane-oxidation in the deep sea. Goldschmidt Conference. Prague.
- Orphan, V.J., Dekas, A.E., Poretsky, R. & Amend, J. (2010). Methane-oxidizing Archaea Fix Nitrogen in Cooperation with Sulfate-reducing Bacteria in Deep-Sea Methane Seeps. Program and Abstracts, AbSciCon10. Houston, TX.
- Rhodes, M.E., Oren, A. & House, C.H. (In Review). Revisiting Dead Sea Haloarchaeal Blooms Through High Throughput Sequencing. Applied and Environmental Microbiology.
- Seth, L., Miteva, V. & Brenchley, J. (2011). Diversity among small-subunit rRNA gene clones obtained from Estisol 240 and Coasol based drilling fluid and post coring chips from a Greenland glacier ice core. Penn State Undergraduate Poster Exhibition.
- Zerkle, A.L., MacAlady, J.L., Jones, D.S. & Farquhar, J. (2011). S isotope investigation of a redox-stratified system dominated by chemotrophic sulfide oxidation. Goldschmidt Meeting. Prague.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
James Ferry
Co-Investigator
Katherine Freeman
Co-Investigator
James Schopf
Co-Investigator
Beth Shapiro
Co-Investigator
Oded Beja
Collaborator
David Bice
Collaborator
Kenneth Broad
Collaborator
Charles Cockell
Collaborator
Sorel Fitz-Gibbon
Collaborator
Kevin McKeegan
Collaborator
Alessandro Montanari
Collaborator
Aharon Oren
Collaborator
John Spear
Collaborator
Roger Summons
Collaborator
Mary Voytek
Collaborator
Paula Welander
Collaborator
Aubrey Zerkle
Collaborator
Jake Bailey
Postdoc
Theresa M. D. Raub
Postdoc
Tim Raub
Postdoc
Jennifer Loveland-Curtze
Research Staff
Vanya Miteva
Research Staff
Irene Schaperdoth
Research Staff
Zhidan Zhang
Research Staff
Heidi Albrecht
Doctoral Student
Daniel Jones
Doctoral Student
Kristine Korzow Richter
Doctoral Student
Rebecca McCauley
Doctoral Student
Moshe (Mathew) Rhodes
Doctoral Student
Abigail M. Green
Graduate Student
Benjamin Harrison
Graduate Student
Norbert Lazar
Graduate Student
Thomas Roberts
Graduate Student
Labdhi Seth
Graduate Student
Anatoliy Kudryavtsev
Unspecified Role
A. Nele Meckler
Unspecified Role
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.3
Effects of extraterrestrial events upon the biosphere
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems