2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Resurrection of an Ancestral Peptidyl Transferase
Project Summary
We have created and test both in silico and in vitro models of an ancestral pepidyl transerase center (PTC). Our most recent in silico and in vitro models contain a significantly reduced 23S rRNA (called a-rRNA-γ, Figure 1), retraining the rRNA that forms and surrounds the PTC. To complete the in silico and in vitro models of the ancestral PTC (a-PTC-γ in silico and a-PTC-γ in vitro), we have combined a-rRNA-γ with peptides derived from the ribosomal proteins. The results here indicate that the ribosome and its components are highly robust in folding and assembly. We have shaved around 2500 nucleotides from the 23S rRNA and the vast majority of amino acids from the protein components, excising the globular domains in toto. Yet, the remaining rRNA and peptides retain the ability to fold and specifically assemble.
Project Progress
The ribosome is a molecular machine responsible for synthesis of coded protein in all living systems. The ribosome is made of a small subunit (SSU) that decodes the messenger RNA and a large subunit (LSU) that catalyzes peptide bond formation.
Some of the RNA and protein components of the ribosome are highly conserved everywhere in extant life, and are considered to be very ancient (1-6). The peptidyl transferase center (PTC), in particular, is thought to be even older than coded protein (7-10). In this view, the PTC emerged from some type of ‘RNA World’ (11-15) participating in the transformation to current biology in which information is transduced from nucleic acids to proteins. In essentially any reasonable model of origin of life and evolution of the ribosome, the PTC appears as one of our most direct biochemical links to the distant evolutionary past and to early life. Understanding the PTC is key to understanding ancient biology.
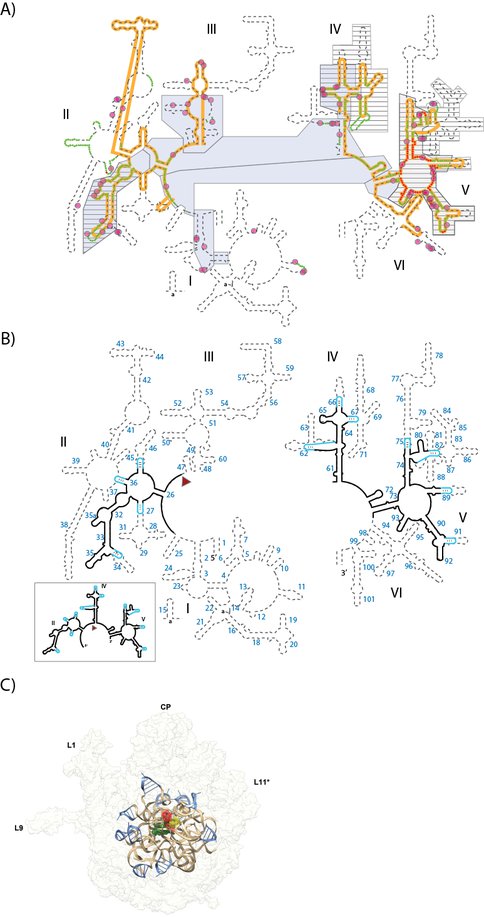
Here we create and test both in silico and in vitro models of an ancestral PTC. Our models incorporate recent proposals (7, 8, 10) that point to portions of Domains V, IV and II of the modern 23S rRNA as elements inherited from an ancestral PTC (Figure 1A). Our in silico and in vitro models contain a significantly reduced 23S rRNA (called a-rRNA-γ, Figure 1B), retraining the rRNA that forms and surrounds the PTC. Using the 3D structure of the T. thermophilus LSU (16) as a template, the 23S rRNA was ‘shaved’ to a rough sphere of around 30 Å in radius (8), centered on the site of peptidyl transfer. Cuts were performed selectively in A-form helical regions, which were capped with stable stem-loops. The result is a single RNA polymer, which is predicted by several proposals of PTC evolution (summarized in Figure 1A), and contains around 20% of the 23S rRNA linked by 11 stem-loops. As illustrated in Figure 1, a-rRNA-γ contains rRNA segments that are (i) are universally conserved in all organisms and organelles (17), (ii) extremely ancient (6, 7) (iii) tightly networked (6), (iv) highly coordinated by magnesium ions (18), and (v) in the central shells of the ribosomal ‘onion’ (8).
To complete the in silico and in vitro models of the ancestral PTC (a-PTC-γ in silico and a-PTC-γ in vitro), we have combined a-rRNA-γ (above) with peptides derived from the ribosomal proteins. Ribosomal proteins do not directly engage in catalysis (19, 20) but do interact extensively with the rRNA that lines the catalytic center. The peptide components of a-PTC-γ are the ‘tail’ segments of ribosomal proteins. Ribosomal proteins L2, L3, L4, L15 and L22 were shaved at the boundary of the sphere defined by a-rRNA-γ. The shaving process did not disrupt protein secondary or tertiary interactions: none of the ribosomal protein tails within a-PTC-γ are globular structures in the intact LSU. The peptide components of a-PTC-γ are called a-rPeptide-L2, a-rPeptide-L3, a-rPeptide-L4, a-rPeptide-L15, and a-rPeptide-L22 (Table 1).
Here, using computation and solution experiments, we test hypotheses that: (i) a-rRNA-γ can assume the canonical LSU secondary structure as shown in Figure 1B, (ii) a-rRNA-γ in association with magnesium can assume LSU-like 3D structure and tertiary interactions as shown in Figure 1C, (iii) a-rRNA-γ forms LSU-like complexes with shaved ribosomal protein segments as shown in Figure 1C, and (iv) a-rRNA-γ in association with these ribosomal protein segments form a functional a-PTC-γ, which is defined by peptidyl transferase catalytic activity. Here a-rPTC-γ was assembled in vitro and in silico, and these hypotheses are tested by various modeling, footprinting, binding and catalytic assays.
Our current models of a-PTC-g are steps in an iterative process that will allow us to test hypotheses related to evolution of translation, including models of RNA folding, assembly, and catalysis. The ultimate goal is to develop biochemically functional ancestral ribosomes. Our current models contain about 500 nucleotides of RNA that thought to predate the coding functionality but not the peptidyl transferase activity of the ribosome. Fragments of ancestral RNA are joined together by stem-loops to form a single RNA polymer called a-rRNA-γ. To assemble a-PTC-g, magnesium ions and six peptides are annealed with a-rRNA-g. The six peptides in a-PTC-gare non-globular fragments of ribosomal proteins representing Fox’s molecular fossils of non-coded precursors of modern proteins (6).
Our fundamental prediction here is that a-PTC-γ in vitro will form the same basic structure, with the same RNA conformation, secondary and tertiary interactions, and the same RNA-protein and RNA-ion interactions, as a-PTC-γ in silico. The 3D structure of a-PTC-γ in silico (Figure 1C) in turn is based on the 3D structure of the extant ribosome (1jj2, T. Thermophilus). The interior of the a-PTC-γ in silico model is very similar to the corresponding region of the extant ribosome. The surface region of a-PTC-γ in silico, in particular the stitching stem-loops, obviously diverge from the extant ribosome.
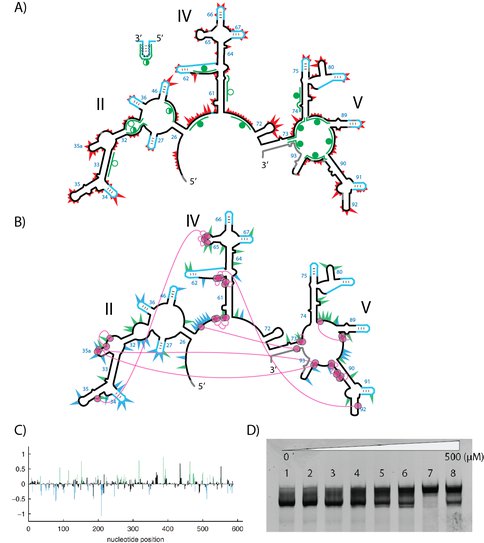
We synthesized the macromolecular components of a-PTC-γ. The gene for a-rRNA-g was assembled by recursive PCR and used as a template in in vitro transcription. The six ancestral peptides were made by solid phase synthesis. The structure and assembly of a-PTC-γ in vitro was probed by a variety of well-established biophysical methods. The results suggest that a-PTC-γ in silico is an excellent predictor of the folding and assembly of a-PTC-γ in vitro. This conclusion is based on (i) RNase H digestion and SHAPE profiles, which support the predicted secondary structure of a-rRNA-γ, (ii) the magnesium-dependence of the SHAPE profile and the magnesium-dependence of gel mobility, which indicate specific magnesium interactions within a-rRNA-g, and magnesium-induced formation of tertiary interactions and (iii) specific binding of ancestral peptides with a-rRNA-g, which is consistent with folding and assembly of a-rRNA-g and the peptides to form a-PTC-g.
The combined results here indicated that the ribosome and its components are highly robust in folding and assembly. We have shaved around 2500 nucleotides from the 23S rRNA and the vast majority of amino acids from the protein components, excising the globular domains in toto. Yet, the remaining rRNA and peptides retain the ability to fold and specifically assemble. It is particularly striking that the ribosomal protein tails, lacking α-helices, β-sheets, and globular domains, retain the ability to interact specifically with a-rRNA-g. This robustness is consistent with the premise that the ribosome is an ancient assembly that evolved in a primitive biological environment, and has survived billions of years of evolution, growing in size and complexity, without major changes in core structure or function. Although the modern LSU has the appearance of a massive monolithic assembly (19), the results here, and elsewhere (21), indicate that it is in fact composed of small RNA and protein elements that retain ancient abilities to fold and assembly independently.
References
1. Woese CR (2000) Interpreting the Universal Phylogenetic Tree. Proc Natl Acad Sci U S A 97:8392-8396.
2. Woese CR (2001) Translation: In Retrospect and Prospect. RNA 7:1055-1067.
3. Fournier GP, Neumann JE, Gogarten JP (2010) Inferring the Ancient History of the Translation Machinery and Genetic Code Via Recapitulation of Ribosomal Subunit Assembly Orders. PLoS One 5:e9437.
4. Williams D, Fournier GP, Lapierre P, Swithers KS, Green AG, Andam CP, Gogarten JP (2011) A Rooted Net of Life. Biol Direct 6:45.
5. Wolf YI, Koonin EV (2007) On the Origin of the Translation System and the Genetic Code in the RNA World by Means of Natural Selection, Exaptation, and Subfunctionalization. Biol Direct 2:14.
6. Fox GE (2010) Origin and Evolution of the Ribosome. Cold Spring Harb Perspect Biol 2:a003483.
7. Bokov K, Steinberg SV (2009) A Hierarchical Model for Evolution of 23S Ribosomal RNA. Nature 457:977-980.
8. Hsiao C, Mohan S, Kalahar BK, Williams LD (2009) Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Mol Biol Evol 26:2415-2425.
9. Belousoff MJ, Davidovich C, Zimmerman E, Caspi Y, Wekselman I, Rozenszajn L, Shapira T, Sade-Falk O, Taha L, Bashan A, Weiss MS, Yonath A (2010) Ancient Machinery Embedded in the Contemporary Ribosome. Biochem Soc Trans 38:422-427.
10. Hury J, Nagaswamy U, Larios-Sanz M, Fox GE (2006) Ribosome Origins: The Relative Age of 23S rRNA Domains. Orig Life Evol Biosph 36:421-429.
11. Rich A (1962) On the Problems of Evolution and Biochemical Information Transfer. Horizons in Biochemistry, eds Kasha M & Pullman B (New York: Academic), pp 103–126.
12. Woese CR (1967) The Genetic Code: The Molecular Basis for Genetic Expression (Harper & Row, N.Y.).
13. Gilbert W (1986) Origin of Life: The RNA World. Nature 319:618-618.
14. Crick FH (1968) The Origin of the Genetic Code. J Mol Biol 38:367-379.
15. Orgel LE (1968) Evolution of the Genetic Apparatus. J Mol Biol 38:381-393.
16. Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science 313:1935-1942.
17. Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, Harvey SC (2002) Modeling a Minimal Ribosome Based on Comparative Sequence Analysis. J Mol Biol 321:215-234.
18. Hsiao C, Williams LD (2009) A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res 37:3134-3142.
19. Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 289:905-920.
20. Noller HF, Hoffarth V, Zimniak L (1992) Unusual Resistance of Peptidyl Transferase to Protein Extraction Procedures. Science 256:1416-1419.
21. Athavale SS, Gossett JJ, Hsiao C, Bowman JC, O’Neill E, E H, Preeprema T, Hud NV, Wartell R, Williams LD (2012) Domain III of the T. Thermophilus 23S rRNA Folds Independently to a near-Native State. RNA:in press.
(A) Various models of ribosomal evolution. The dashed line shows the canonical secondary structure of the T. thermophilus 23S rRNA. Secondary structural domains are indicated by Roman numerals. The red and green highlights show the two inner shells of the ribosomal onion, which marks the rRNA that is in closest proximity, in three dimensions, to the site of peptidyl transfer. The gray boxes are ancient according to the ‘A-minor’ method of Steinberg. The hashed boxes (with black horizontal lines) are ancient according the ‘Foreign Graduate Student’ method of Fox. First shell magnesium-phosphate interactions are indicated by magenta circles. The orange line shows the universally conserved portions of the 23S rRNA, in bacteria, archaea, eukarya and in eukaryotic organelles, as determined by Gutell and Harvey. (B) Secondary structure of the proposed ancestral rRNA (a-rRNA-γ) which is derived from a consensus of models of rRNA evolution. The rRNA fragments of the model are taken directly from the T. thermophilus 23S rRNA and are indicated by black lines in the secondary structure. Stable stem loops that stitch these rRNA fragments together are blue. Helix numbers are indicated. The predicted secondary structure of the a-rRNA-γ alone is highlighted in the outbox. (C) a-PTC-γ in silico. A three-dimensional model of a-PTC-γ, composed of a-rRNA-γ and five peptides (fragments of ribosomal proteins L2, L3, L4, L15 and L22). The core of the model is identical to the central core of the T. Thermophilus LSU. a-rRNA-γ is shown in brown ribbon representation, the stitching stem-loops are shown in blue, and the peptides are shown in green spheres. For references A-site (yellow), P-site (red) substrates are shown in the figure, but are not a component of a-PTC-γ in silico. The entire 23S rRNA is indicated in light gray ribbon.
Probing the secondary and tertiary structure of a-rRNA-γ. (A) SHAPE and RNase analysis. Red triangles mark the SHAPE reactivities in 250 mM Na+, mapped onto the predicted secondary structure of a-rRNA-γ. Larger triangles indicates higher SHAPE reactivity. RNase H DNA probes are marked green lines. The circle indicates the degree of digestion of RNA by RNase H: filled circles (more than 70%), half-filled circle (between 25% and 75%), and empty circles (less than 25%). (B) Highly dispersed effects of magnesium on SHAPE reactivity suggest formation of tertiary structure. Green triangles show the greatest increases in SHAPE reactivity upon addition of magnesium. Blue triangles show the greatest decreases in reactivity. First-shell magnesium-phosphate interactions observed in the T. thermophilus LSU (PDB entry: 2J00 and 2J01) are mapped onto the predicted secondary structure of a-rRNA-γ. Magenta circles indicate first-shell Mg-OP (magnesium-phosphate) interactions. The lines indicate PO-Mg-OP linkages. In both panels gray shading indicates regions of rRNA where SHAPE data were unobtainable. (C) [SHAPE reactivity of the a-RNA-γ in the absence of Mg2+. Blue and green are reactivities changes (> 0.3) in the addition of Mg2+ ions. (D) The effect of magnesium on a-rRNA-γ gel mobility suggest formation of tertiary structure. Shown here is a-PTC-γ annealed in varying magnesium concentrations, separated on a 5% native acrylamide gel. The Mg2+ concentrations are: Lane 1: no Mg2+; Lane 2: 1 μM; Lane 3: 12.5 μM; Lane 4: 25 μM, Lane 5: 50 μM, Lane 6: 100 μM, Lane 7: 250 μM, Lane 8: 500 μM.
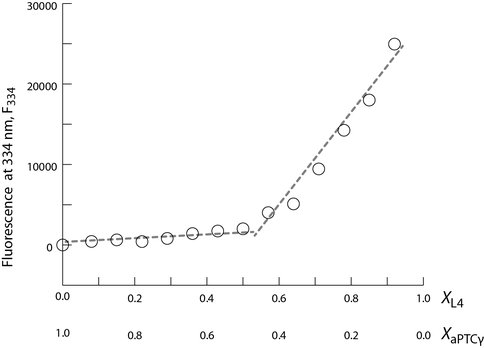
a-rRNA-γ and a-rPeptide-L4 are assembly competent and form a complex with 1:1 stoichiometry. The circles denote the fluorescence intensity of the tryptophan residues of a-rPeptide-L4 measured at 334 nm in a continuous variation experiment with a-rRNA-γ. The discontinuity at a equivalent mole fractions indicates a complex with 1:1 stoichiometry.
Publications
-
Dasgupta, I., Gao, X., & Fox, G. E. (2011). Structural properties of DNA oligomers containing (GACX)n and (GAXC)n tandem repeats. Biopolymers, 97(3), 155–164. doi:10.1002/bip.21719
-
Fox, G. E., Tran, Q., & Yonath, A. (2012). An Exit Cavity Was Crucial to the Polymerase Activity of the Early Ribosome. Astrobiology, 12(1), 57–60. doi:10.1089/ast.2011.0692
-
Liu, Y., Stepanov, V. G., Strych, U., Willson, R. C., Jackson, G. W., & Fox, G. E. (2010). DNAzyme-mediated recovery of small recombinant RNAs from a 5S rRNA-derived chimera expressed in Escherichia coli. BMC Biotechnol, 10(1), 85. doi:10.1186/1472-6750-10-85
-
Lu, Q., & Fox, G. E. (2011). Resurrection of an ancestral 5S rRNA. BMC Evolutionary Biology, 11(1), 218. doi:10.1186/1471-2148-11-218
-
Nayar, M., & Fox, G. E. (2011). Defining 5S rRNA Structure Space: Point Mutation Data Can Be Used to Predict the Phenotype of Multichange Variants. Molecular Biology and Evolution, 28(9), 2629–2636. doi:10.1093/molbev/msr090
-
Updegrove, T. B., & Wartell, R. M. (2011). The influence of Escherichia coli Hfq mutations on RNA binding and sRNA•mRNA duplex formation in rpoS riboregulation. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms, 1809(10), 532–540. doi:10.1016/j.bbagrm.2011.08.006
-
Updegrove, T. B., Correia, J. J., Chen, Y., Terry, C., & Wartell, R. M. (2011). The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA, 17(3), 489–500. doi:10.1261/rna.2452111
-
Updegrove, T. B., Correia, J. J., Galletto, R., Bujalowski, W., & Wartell, R. M. (2010). E. coli DNA associated with isolated Hfq interacts with Hfq’s distal surface and C-terminal domain. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms, 1799(8), 588–596. doi:10.1016/j.bbagrm.2010.06.007
-
Zhao, Q., Huang, H-C., Nagaswamy, U., Xia, Y., Gao, X., & Fox, G. E. (2012). UNAC tetraloops: to what extent do they mimic GNRA tetraloops?. Biopolymers, 97(8), 617–628. doi:10.1002/bip.22049
- Fox, G.E. (2011). The evolutionary history of the ribosome and its relevance to the search for life elsewhere in the universe. SPIE Conference 8152; Instruments, Methods and Missions for Astrobiology XIV.
- Jain, K., Updegrove, T.B. & Wartell, R.M. (2011). Thermodynamics of sRNA-mRNA interactions and the role of Hfq. In: Sheardy, R. (Eds.). Frontiers in Nucleic Acids (ACS Symposium Series).
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Shreyas Athavale
Postdoc
Chiaolong Hsiao
Postdoc
Emmanuel Tannenbaum
Postdoc
Madhan Tirumalai
Postdoc
Jessica Bowman
Research Staff
Eric O'Neill
Research Staff
Josh Canzoneri
Graduate Student
Jared Gossett
Graduate Student
Timothy Lenz
Graduate Student
Lively Lie
Graduate Student
Thanawadee Preeprem
Graduate Student
Quyen Tran
Graduate Student
Arren Washington
Graduate Student
Amy Boudreau
Undergraduate Student
Kaylee Goss
Undergraduate Student
Suk Hahm
Undergraduate Student
Jason Murray
Undergraduate Student
Jessica Peters
Undergraduate Student
Joshua Raimist
Undergraduate Student
Catherine Trippe
Undergraduate Student
Andrew Warren
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.2
Production of complex life.