2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Ironing Out the RNA World
Project Summary
Life originated during the early Archean, which was characterized by a non-oxidative atmosphere and abundant soluble Fe2+. Current theories on the origin of life focus on RNA-based genetic and metabolic systems. Here we show, by theory and experiment, that critical roles of Mg2+ in extant RNA folding and function can be better served by Fe2+ in the absence of oxygen. The results of our high-level quantum mechanical calculations show that the geometry of coordination of Fe2+ by RNA phosphates is similar to that of Mg2+. The conformation of Tetrahymena Group I intron P4-P6 domain is conserved between complexes of Fe2+ and Mg2+. Additionally, a ribozyme obtained previously by in vitro selection in the presence of Mg2+ and a natural ribozyme both have significantly greater catalytic competence in the presence of Fe2+ than in Mg2+. The combined biochemical and paleogeological data are consistent with an RNA-Fe2+ world that could have supported an array of RNA structures and catalytic functions far more diverse that of an RNA-Mg2+ world.
Project Progress
For at least the last half-billion years, the earth’s biosphere has been rich in free oxygen: a product of photosynthesis. With the rise in free oxygen much of the biologically available iron (Fe2+) was oxidized and deposited in banded iron formations (BIFs) (1). With photosynthesis, the early earth’s abundant Fe2+ was oxidized and sequestered to the extent that current biomass and species diversity in many ecosystems is limited by Fe2+ availability (2).
Living systems are dependent on and must acquire and utilize iron. Yet, iron combined with oxygen is toxic and expensive to manage (3, 4). The concentration of iron in cells is on the order of 100 μM, with iron largely constrained to heme, iron-sulfur clusters, and di-iron or mono-iron centers, or to transporters, carriers, exporters, and concentrators such as ferritins (5). Because the solubility of ferric iron in water or plasma is very low (10-18 M), cells must combat a massive internal to external concentration gradient.
It was not always thus. Life originated and thrived for billions of years on earth in an anoxic environment in which iron was soluble, abundant, and non-toxic. BIF iron was once soluble and is seen by isotopic variations to have participated in ancient biological processes (6). The transition from benign and abundant iron to scarce and toxic iron caused a slow but dramatic shift in biology that required radical transformations in biochemical mechanisms and metabolic pathways.
It is believed that life originated with RNA-based genetic and metabolic systems, i.e. the RNA world (7). Questions naturally arise as to whether Fe2+ interacted extensively with RNA during the long anoxic period before photosynthesis. Could Fe2+ have facilitated ancient RNA folding and catalysis? Was Fe2+ replaced by Mg2+ with the rise of oxygen and an increase in Mg2+ availability? Are the RNA-Fe2+ complexes observed in extant biology (8) a type of molecular fossil from the RNA world? Here we address the structural and catalytic relationship between RNA and Fe2+ in the absence of oxygen. We use quantum mechanical calculations, chemical footprinting and ribozyme assays to demonstrate that Fe2+ can readily substitute for Mg2+ in RNA folding and catalysis. The relationship of Fe2+ with RNA could provide a missing link between the geological record and the RNA world.
Theory shows that RNA conformation is conserved in the presence of Fe2+ or Mg2+. The results of our high-level quantum mechanical (QM) calculations show that the geometry of coordination of Fe2+ by RNA phosphates is similar to that of Mg2+. The RNA-Mg2+ complex characterized here by computation represents the most frequent mode of multidentate coordination of Mg2+ in large RNAs, where a Mg2+ ion is coordinated by oxyanions of adjacent nucleotides (9, 10) (Figure 1A). One observes twenty-five of these RNA-Mg2+ clamps in the Haloarcula marisortui large ribosomal subunit (11), two in the P4-P6 domain of the Tetrahymena thermophila Group 1 intron (12), one in a self-splicing group II intron from Oceanobacillus iheyensis (13), and one in the L1 ribozyme ligase (14). The folding and function of each of these RNAs is Mg2+ dependent.
The conformations of RNA clamps with Mg2+ or Fe2+ determined by high-level theory are virtually indistinguishable. The metal-oxygen distances and angles are nearly identical (Figures 1B &1C). The QM calculations do indicate some subtle differences, however. The energies of complex formation favor Fe2+ over Mg2+ by 1.4 kcal/mol, as revealed by Natural Bond Order analysis (15). More charge is transferred from phosphate to Fe2+ (0.43 e-) than from phosphate to Mg2+ (0.29 e-), suggesting that compared to Mg2+, Fe2+ better activates the phosphorous of RNA to nucleophilic attack. These differences are attributable to the greater accessibility of Fe2+ d-orbitals. The similarities of the interactions of Fe2+ or Mg2+ in complexes with RNA are consistent with the interchangeability of Fe2+ and Mg2+ in phosphate minerals such as lazulite [(Mg2+,Fe2+)(Al3+)2(PO43-)2(OH-)2].
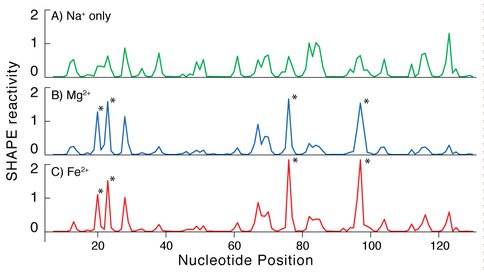
Chemical probing confirms that RNA conformation is conserved in the presence of Fe2+ or Mg2+. The conservation of RNA conformation in association with Fe2+ or Mg2+ predicted in silico is observed in vitro. In solution, RNA conformation can be assayed by chemical footprinting using SHAPE (16) (Selective 2’-Hydroxyl Acylation analyzed by Primer Extension). These chemical footprints are known to be dependent on Mg2+ concentration (16, 17). We used SHAPE to compare RNA conformation with Fe2+ to that with Mg2+. We observe that in the absence of oxygen, the addition of Fe2+ or Mg2+ causes pronounced and nearly identical changes in the SHAPE reactivity of the Tetrahymena Group I intron P4-P6 domain (Figure 2). These results suggest that the ‘magnesium ion core’ of the P4-P6 domain can be recapitulated by Fe2+ in the absence of oxygen.
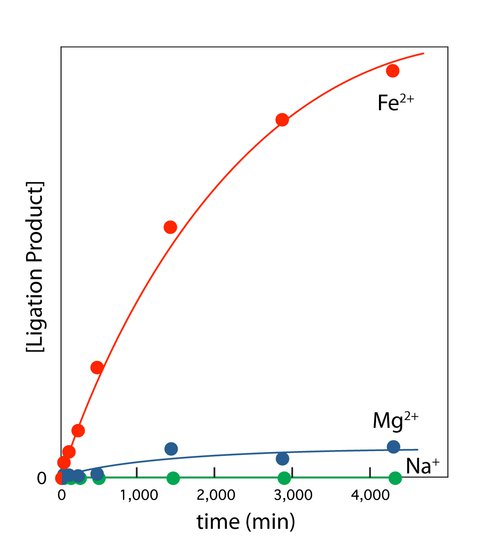
Ribozyme activity is enhanced by Fe2+ compared to Mg2+. We investigated the catalytic activity of the L1 ribozyme ligase in the presence of Mg2+ and Fe2+ (in the absence of oxygen). This ligase catalyzes formation of a phosphodiester linkage during which the 3′-hydroxyl group of an RNA substrate attacks the α-phosphorus of the ribozyme 5′-triphosphate (14). This ribozyme was obtained by Robertson and Ellington by in vitro selection in the presence of high [Mg2+] (60 mM), and has been described as Mg2+-dependent (14). The initial rate of ligation is over 10-fold higher in 100 μM Fe2+ than in 100 μM Mg2+ (Figure 3). The Fe2+ advantage holds for essentially any equimolar concentration of the two divalent cations. Achieving an equivalent rate of reaction requires around a 10-fold greater [Mg2+] than [Fe2+]. As expected, this ligase is inactive in Na+ alone. To explore whether enhancement of ribozyme activity by Fe2+ is a general effect, we also examined the hammerhead ribozyme. Fe2+, compared to Mg2+, causes a significant enhancement of activity of that natural ribozyme.
Some of the first studies of RNA folding showed that magnesium is required for formation of tertiary structure of tRNA (18). It is now known that magnesium plays specific roles in folding of essentially all large RNAs (19, 20). Magnesium shares a distinctive geometric and electronic relationship with phosphate groups. The ionic radius of Mg2+ is small, the charge density is high, the coordination geometry is strictly octahedral, the preferred ligands are charged or neutral oxygens, and the hydration enthalpy is large (21-23). In comparison with group I ions, calcium, or polyamines (24), magnesium has a greater affinity for phosphate oxygens, and can bind to them with well-defined geometry.
Here we provide support for a model of the RNA world, extending into the early protein/DNA/RNA world, in which Fe2+, not Mg2+, was the predominant divalent partner for RNA. The conformations of RNA clamps with Mg2+ or Fe2+ determined by high-level quantum mechanical calculations are virtually indistinguishable. The metal-oxygen distances and angles are nearly identical (Figures 1B &1C). Facile substitution of Mg2+ by Fe2+ is confirmed by chemical probing experiments that show that changes in SHAPE reactivity of RNA induced by association with Mg2+ or Fe2+ are very similar (Figures 2B and 2C). And finally, at equimolar concentrations of Mg2+ or Fe2+, the initial rate of ligation observed for the L1 ribozyme ligase is over 10-fold higher with Fe2+ than Mg2+ (Figure 3).
The RNA world is hypothesized to have occurred during the early Archean eon. During this time it is thought that Mg2+ marine concentration was attenuated by submarine hydrothermal systems associated with higher heat flow (25, 26) and more vigorous seafloor spreading (27, 28), and by reduced Mg2+ delivery to the oceans by smaller continental landmasses. During the early Archean eon, Fe2+ would have been soluble and non-toxic because the environment was anoxic. The [Fe2+]marine in the early Archean is largely circumscribed in current models by PCO2 in the atmosphere (Fe2+ precipitates as siderite: Fe2+CO3-). Early earth PCO2 is inferred using estimates of greenhouse effects, sun luminosity, earth albedo and temperatures required to maintain liquid oceans. Recent indications of a low albedo early earth dispense with requirements for high greenhouse PCO2 (29). If so, [Fe2+]marine could have been as high as 100-1000 mM (30, 31).
The combined biochemical and geological data are consistent with an RNA-Fe2+ world. It is nearly inconceivable that RNA in the early Archean eon escaped extensive interaction with Fe2+. Our observations of Fe2+-mediated RNA folding and catalysis, in combination with the paleogeological models, suggest that RNA functioned and evolved in association with Fe2+. This RNA-Fe2+ world could have supported an array of RNA structures and catalytic functions far more diverse than that of a RNA-Mg2+ world. Complexes of RNA with Fe2+ offer the prospect of a rich redox chemistry for ancient ribozymes.
References
1. Klein C (2005) Some Precambrian Banded Iron-Formations (BIFs) from around the World: Their Age, Geologic Setting, Mineralogy, Metamorphism, Geochemistry, and Origin. Am Mineral 90:1473-1499.
2. Sunda WG, Huntsman SA (1995) Iron Uptake and Growth Limitation in Oceanic and Coastal Phytoplankton. Mar Chem 50:189-206.
3. Kell DB (2009) Iron Behaving Badly: Inappropriate Iron Chelation as a Major Contributor to the Aetiology of Vascular and Other Progressive Inflammatory and Degenerative Diseases. BMC Med Genomics 2:2.
4. Chiancone E, Ceci P, Ilari A, Ribacchi F, Stefanini S (2004) Iron and Proteins for Iron Storage and Detoxification. BioMetals 17:197-202.
5. Theil EC, Goss DJ (2009) Living with Iron (and Oxygen): Questions and Answers About Iron Homeostasis. Chem Rev 109:4568-4579.
6. Johnson CM, Beard BL, Roden EE (2008) The Iron Isotope Fingerprints of Redox and Biogeochemical Cycling in Modern and Ancient Earth. Annual Review of Earth and Planetary Sciences 36:457-493.
7. Anonymous (2011) RNA Worlds: From Life’s Origins to Diversity in Gene Regulation. eds Atkins JF, Gesteland RF, & Cech TR (Cold Spring Harbor Laboratory Press).
8. Goss DJ, Theil EC (2011) Iron Responsive mRNAs: A Family of Fe(2+) Sensitive Riboregulators. Acc Chem Res.
9. Petrov AS, Bowman JC, Harvey SC, Williams LD (2011) Bidentate RNA-Magnesium Clamps: On the Origin of the Special Role of Magnesium in RNA Folding. RNA 17:291-297.
10. Hsiao C, Williams LD (2009) A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res 37:3134-3142.
11. Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 289:905-920.
12. Cate JH, Hanna RL, Doudna JA (1997) A Magnesium Ion Core at the Heart of a Ribozyme Domain. Nat Struct Biol 4:553-558.
13. Toor N, Keating KS, Taylor SD, Pyle AM (2008) Crystal Structure of a Self-Spliced Group II Intron. Science 320:77-82.
14. Robertson MP, Scott WG (2007) The Structural Basis of Ribozyme-Catalyzed RNA Assembly. Science 315:1549-1553.
15. Glendening ED, Streitwieser A (1994) Natural Energy Decomposition Analysis – an Energy Partitioning Procedure for Molecular-Interactions with Application to Weak Hydrogen-Bonding, Strong Ionic, and Moderate Donor-Acceptor Interactions. J Chem Phys 100:2900-2909.
16. Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM (2005) RNA Structure Analysis at Single Nucleotide Resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J Am Chem Soc 127:4223-4231.
17. Mortimer SA, Weeks KM (2008) Time-Resolved RNA SHAPE Chemistry. J Am Chem Soc 130:16178-16180.
18. Stein A, Crothers DM (1976) Conformational Changes of Transfer RNA. The Role of Magnesium(II) Equilibrium Binding of Magnesium(II) by Escherichia Coli tRNAfMet. Biochemistry 15:160-168.
19. Draper DE (2008) RNA Folding: Thermodynamic and Molecular Descriptions of the Roles of Ions. Biophys J 95:5489-5495.
20. Auffinger P, Grover N, Westhof E (2011) Metal Ion Binding to RNA [Review]. Met Ions Life Sci 9:1-35.
21. Brown ID (1992) Chemical and Steric Constraints in Inorganic Solids. Acta Crystallogr Sect B 48:553-572.
22. Rashin AA, Honig B (1985) Reevaluation of the Born Model of Ion Hydration. J Phys Chem 89:5588-5593.
23. Bock CW, Markham GD, Katz AK, Glusker JP (2006) The Arrangement of First- and Second-Shell Water Molecules around Metal Ions: Effects of Charge and Size. Theor Chem Acc 115:100-112.
24. Feuerstein BG, Williams LD, Basu HS, Marton LJ (1991) Implications and Concepts of Polyamine-Nucleic Acid Interactions. J Cell Biochemistry 46:37-47.
25. Condie KC (2007) The Distribution of Paleoarchean Crust. Earth’s Oldest Rocks, The Distribution of Paleoarchean Crust, eds Van Kranendonk MJ, Smithies RH, & Bennett VC (Elsevier Publishers, Amsterdam).
26. Tajika E, Matsui T (1992) Evolution of Terrestrial Proto-CO2 Atmosphere Coupled with Thermal History of the Earth. Earth and Planetary Science Letters 113:251-266.
27. Condie KC, Kroner A (2008) When Did Plate Tectonics Begin? Evidence from the Geologic Record. Geological Society of America Special Papers, eds Condie KC & Pease V), Vol 440, pp 281-294.
28. Davies GF (1992) On the Emergence of Plate-Tectonics. Geology 20:963-966.
29. Rosing MT, Bird DK, Sleep NH, Bjerrum CJ (2010) No Climate Paradox under the Faint Early Sun. Nature 464:744-747.
30. Holland H (1984) The Chemical Evolution of the Atmosphere and Oceans (Princeton University Press, Princeton, New Jersey) p 582.
31. Anbar AD (2008) Oceans. Elements and Evolution. Science 322:1481-1483.
Conformations of RNA-Mg2+ and RNA-Fe2+ clamps are nearly identical. A) RNA-Mg2+ clamp from the L1 ribozyme ligase (PDB 2OIU). B) RNA-Mg2+ clamp optimized by high level QM calculations. C) An optimized RNA-Fe2+ clamp. Each cation (Mg2+ or Fe2+) is hexacoordinate. Mg2+ is shown as a yellow sphere and Fe2+ is shown as a green sphere. Water molecules are omitted from the images for clarity. Distances are in Å.
Addition of Mg2+ or Fe2+ causes the same changes in the SHAPE reactivity of the P4-P6 domain of the Tetrahymena Group 1 intron. A) Shape profile in presence of 250 mM NaCl and no divalent cations. B) The addition of Mg2+ increases the reactivity at the sites indicated with the asterisks and decreases reactivity at other sites. This reaction contains 2.5 mM Mg2+ and 250 mM NaCl. C) The addition of Fe2+ causes the same changes in SHAPE reactivity as Mg2+. This reaction contains 2.5 mM Fe2+ and 250 mM NaCl.
L1 ribozyme ligase activity is enhanced by Fe2+ compared to Mg2+. L1 ribozyme ligase reactions were performed at room temperature and 250 mM Na+ in 100 mM [Fe2+] or 100 mM [Mg2+]. The reaction components were first annealed in 50 mM HEPES, pH 8.0, 200 mM sodium acetate by incubating at 90°C for 3 min and cooling to room temperature over 30 min. The ligation reaction was initiated by adding the appropriate cation salt. The Na+ only reaction gave no product. Reaction progress was monitored by gel electrophoresis.
Publications
- Athavale, S.S., Petrov, A.S., Hsiao, C., Watkins, J.D., Prickett, C., Gossett, J.J., Bowman, J.C., O’Neill, E., Bernier, C., Hud, N.V., Wartell, R. & Williams, L.D. (2012, In Review). Ironing out the RNA World.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Shreyas Athavale
Postdoc
Chiaolong Hsiao
Postdoc
Anton Petrov
Postdoc
J. Derrick Watkins
Postdoc
Jessica Bowman
Research Staff
Eric O'Neill
Research Staff
Caitlin Prickett
Research Staff
Chad Bernier
Graduate Student
Jared Gossett
Graduate Student
Lively Lie
Graduate Student
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.