2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Domain III of the 23S rRNA: An Independent Domain
Project Summary
The three-dimensional structure of the ribosomal large subunit (LSU) reveals a single morphological element, although the 23S rRNA is contained in six secondary structure domains. Based upon maps of inter- and intra-domain interactions and proposed evolutionary pathways of development, we hypothesize that Domain III is a truly independent structural domain that can fold to a near-native state in the absence of the remainder of the LSU. Domain III is primarily stabilized by intra-domain interactions, negligibly perturbed by inter-domain interactions, and is not penetrated by proteins or other rRNA. We have probed the structure of Domain III rRNA alone and when contained within the intact 23S rRNA using SHAPE (selective 2’-hydroxyl acylation analyzed by primer extension), in the absence and presence of magnesium. The combined results support the hypothesis that Domain III alone folds to a near-native state with secondary structure, intra-domain tertiary interactions and inter-domain interactions that are independent of whether or not it is embedded in the intact 23S rRNA or within the LSU. The data presented support previous suggestions that Domain III was added relatively late in ribosomal evolution.
Project Progress
The ribosome is made of a small subunit (SSU) and a large subunit (LSU). The SSU in bacteria and archaea contains a single RNA molecule, the 16S rRNA. Phylogenic studies by Woese and coworkers (1) revealed three major and one minor secondary structural domains (2° domains) of the 16S rRNA. These 2° domains are segregated into independent and autonomous three-dimensional domains (3D domains) in the assembled SSU. Each 2° domain of the 16S rRNA folds and assembles with the appropriate ribosomal proteins into a 3D domain, independent of other 2° domains (2-4). One 3D domain is called the head and others are called the body and the platform (5, 6). The head, body and platform domains of the SSU have direct functional significance, moving independently during translation (7). These 3D domains may also have evolutionary significance. The domain is the evolutionary unit of protein evolution (8, 9). Protein domains are modular units that are combined and recombined over evolution to achieve various functions. It is conceivable that the 3D domains of the SSU played analogous evolutionary roles, but on a more ancient timeframe. If so, the 3D domains of the SSU may have been recruited to the ribosome, from prior functional roles.
The LSU in bacteria and archaea is made up of a 23S rRNA and a much smaller 5S rRNA. The 23S rRNA contains six 2° domains (10) (Figure 1A). Although these 2° domains are well-defined in the secondary structure, in three dimensions the LSU appears monolithic (11-13). It has been suggested that, unlike in the SSU, the 2° domains in the LSU do not correspond to 3D domains.
Questions naturally arise as to whether the architectures and early evolution of the SSU and the LSU are fundamentally different, and if so, why? Do isolated 2° domains of the 16S rRNA but not the 23S rRNA act as 3D domains and fold to near-native 3D structures? How are the 2° domains of the 16S and 23S rRNAs related to 3D structure, function and evolution of the ribosome?
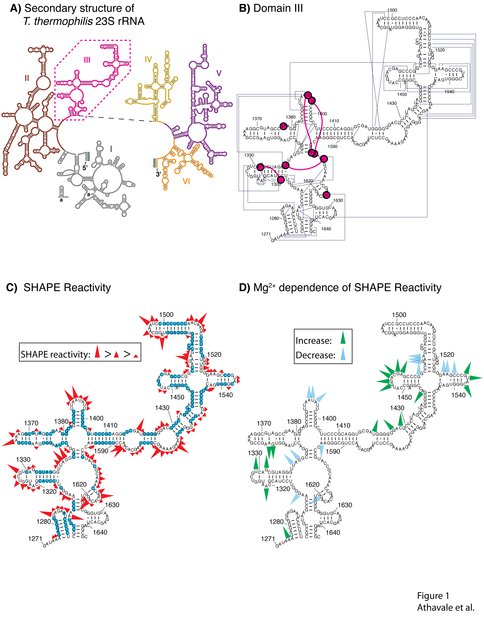
Here we experimentally probe the domain structure of the LSU. We show that one isolated 2° domain of the 23S rRNA can fold to a near-native state in absence of the remainder of the LSU, and appears to be a true 3D domain. Our focus here is Domain III of the _T. thermophilus 23S rRNA (Figure 1B), which is described by Thirumalai and coworkers (14) as compact and slightly prolate. We use SHAPE (15, 16) to demonstrate that Domain III excised from the 23S rRNA (Domain IIIalone) folds in a magnesium-dependent fashion to the same basic state as when it is embedded in the intact 23S rRNA (Domain III23S). In this near-native state of Domain III, surface residues appear to be poised with the correct geometry for the inter-domain rRNA-rRNA interactions observed in the structure of the LSU (PDB entry 2J01) (17). Our results are consistent with the structure of Domain III within the LSU where Domain III is compact, and its interactions with other ribosomal components are restricted to its surface (Figures 2 and 3).
The domain structures of rRNAs have profound implications for folding and function of the ribosome, and early evolution of life. In contrast to the SSU, it has been proposed that the 2° domains of the LSU (Figure 1A) are melded into a single monolithic unit (11-13). LSU 2° domains are thought to be so highly intertwined and interconnected that they lack distinct structural and functional significance and are not true 3D domains.
Considering the extensive network of intra-domain tertiary interactions of Domain III (Figure 1B) and its isolation from the inter-domain network of molecular interactions within the LSU, we hypothesize that Domain III is a true 3D domain. By contrast, domain V, which contains the Peptidyl Transferase Center, is extensively networked with other 2° domains. Domain V makes 24 inter-domain A-minor interactions (18). Additionally, Domain V makes six inter-domain magnesium-mediated phosphate-phosphate linkages (19). Domain III only makes six A-minor interactions and single magnesium-mediated phosphate-phosphate linkage with other 2° domains.
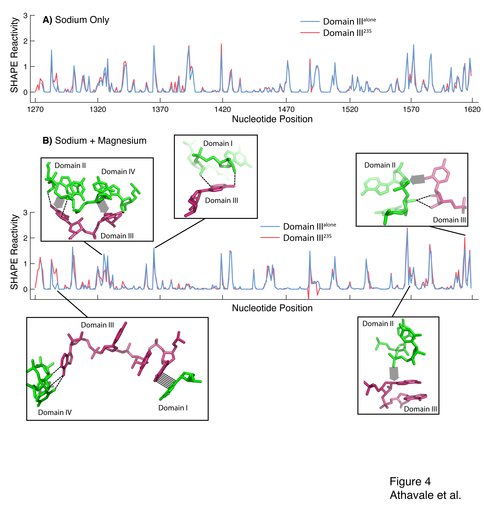
We present data indicating that Domain IIIalone adopts a secondary structure that is the same as Domain III23S (Figures 1C and 4A). The addition of magnesium facilitates folding to a near-native state of both Domain IIIalone and Domain III23S, with the formation of intra-domain tertiary interactions (Figures 1D and 4B). The disruption of inter-domain interactions of Domain III is reflected in the subtle but observable changes in SHAPE reactivity when Domain III is excised from the 23S rRNA (i.e., when Domain III23S is converted to Domain IIIalone, Figure 4B). The mapping of these changes in SHAPE reactivity to regions of inter-domain interactions is evidence that Domain IIIalone and Domain III23S fold to near-native states. This interpretation is supported by the previous observation that Domain IIIalone interacts specifically with ribosomal protein L23 (20).
In sum it appears that, like the SSU, the LSU also contains some elements of a 3D domain-based architecture, in spite of its monolithic appearance. At least some 2° domains of the 23S rRNA (Domain III) autonomously fold to near-native states apart from the rest of the LSU. Consequently, at least some LSU 2° domains may have played roles similar to SSU 2° domains during the evolutionary development of the ribosome. Previous support for the importance of 3D domains of the LSU is found in the demonstration that Domain I alone is highly structured (21). Further, Garret and co-workers have demonstrated that isolated domains of the 23S rRNA are able to form the correct secondary structure and bind to specific ribosomal proteins (20-22).
Evolutionary Implications of the Domain Structure of the Domain III. The ribosome in its present form was well established at the emergence of the last universal common ancestor of life (LUCA) (18, 23-28). There is a consensus that some parts of the ribosome are even older than LUCA, predating the protein world. Parts of Domain V of the 23S rRNA is believed to be among the most ancient parts of the ribosome (18, 23-28) while Domain III is thought to be a more recent addition (29). The data presented here support the hypothesis that Domain III was added as an intact entity to the ancestral ribosome – assuming the 3D domain is a unit of ribosomal evolution. This evolutionary model is consistent with the absence of Domain III from certain mitochondrial rRNAs, such as that of Trypanosoma brucei (30). Ribosomes in which Domain III is absent may have had this domain deleted by relatively recent evolutionary processes within the mitochondrion, but presumably retained functionality with the assistance of proteins.
References
1. Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, Noller HF (1980) Secondary Structure Model for Bacterial 16S Ribosomal RNA: Phylogenetic, Enzymatic and Chemical Evidence. Nucleic Acids Res 8:2275-2293.
2. Samaha RR, O’Brien B, O’Brien TW, Noller HF (1994) Independent in Vitro Assembly of a Ribonucleoprotein Particle Containing the 3’ Domain of 16S rRNA. Proc Natl Acad Sci U S A 91:7884-7888.
3. Agalarov SC, Selivanova OM, Zheleznyakova EN, Zheleznaya LA, Matvienko NI, Spirin AS (1999) Independent in Vitro Assembly of All Three Major Morphological Parts of the 30S Ribosomal Subunit of Thermus Thermophilus. Eur J Biochem 266:533-537.
4. Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J (1993) Chemical Evidence for Domain Assembly of the Escherichia Coli 30S Ribosome. FASEB J 7:177-180.
5. Brimacombe R, Maly P, Zwieb C (1983) The Structure of Ribosomal RNA and Its Organization Relative to Ribosomal Protein. Prog Nucleic Acid Res Mol Biol 28:1-48.
6. Wimberly BT, Brodersen DE, Clemons WM, Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S Ribosomal Subunit. Nature 407:327-339.
7. Noller HF (2005) RNA Structure: Reading the Ribosome. Science 309:1508-1514.
8. Campbell ID, Downing AK (1994) Building Protein Structure and Function from Modular Units. Trends Biotechnol 12:168-172.
9. Fong JH, Geer LY, Panchenko AR, Bryant SH (2007) Modeling the Evolution of Protein Domain Architectures Using Maximum Parsimony. J Mol Biol 366:307-315.
10. Noller HF, Kop J, Wheaton V, Brosius J, Gutell RR, Kopylov AM, Dohme F, Herr W, Stahl DA, Gupta R, Woese CR (1981) Secondary Structure Model for 23S Ribosomal RNA. Nucleic Acids Res 9:6167-6189.
11. Tumminia SJ, Hellmann W, Wall JS, Boublik M (1994) Visualization of Protein-Nucleic Acid Interactions Involved in the in Vitro Assembly of the Escherichia Coli 50S Ribosomal Subunit. J Mol Biol 235:1239-1250.
12. Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 289:905-920.
13. Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal Structure of the Ribosome at 5.5 a Resolution. Science 292:883-896.
14. Hyeon C, Dima RI, Thirumalai D (2006) Size, Shape, and Flexibility of RNA Structures. J Chem Phys 125:194905.
15. Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM (2005) RNA Structure Analysis at Single Nucleotide Resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J Am Chem Soc 127:4223-4231.
16. Wilkinson KA, Merino EJ, Weeks KM (2005) RNA SHAPE Chemistry Reveals Nonhierarchical Interactions Dominate Equilibrium Structural Transitions in tRNA(Asp) Transcripts. J Am Chem Soc 127:4659-4667.
17. Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science 313:1935-1942.
18. Bokov K, Steinberg SV (2009) A Hierarchical Model for Evolution of 23S Ribosomal RNA. Nature 457:977-980.
19. Hsiao C, Williams LD (2009) A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res 37:3134-3142.
20. Ostergaard P, Phan H, Johansen LB, Egebjerg J, Ostergaard L, Porse BT, Garrett RA (1998) Assembly of Proteins and 5S rRNA to Transcripts of the Major Structural Domains of 23S rRNA. J Mol Biol 284:227-240.
21. Egebjerg J, Leffers H, A. C, Andersen H, Garrett RA (1987) Structure and Accessibility of Domain I of Escherichia Coli 23S RNA in Free RNA, in the L24-RNA Complex and in 50S Subunits. Implications for Ribosomal Assembly. J Mol Biol 196:125-136.
22. Leffers H, Egebjerg J, Andersen A, Christensen T, Garrett RA (1988) Domain Vi of Escherichia Coli 23S Ribosomal RNA. Structure, Assembly and Function. J Mol Biol 204:507-522.
23. Belousoff MJ, Davidovich C, Zimmerman E, Caspi Y, Wekselman I, Rozenszajn L, Shapira T, Sade-Falk O, Taha L, Bashan A, Weiss MS, Yonath A (2010) Ancient Machinery Embedded in the Contemporary Ribosome. Biochem Soc Trans 38:422-427.
24. Hsiao C, Mohan S, Kalahar BK, Williams LD (2009) Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Mol Biol Evol 26:2415-2425.
25. Smith TF, Lee JC, Gutell RR, Hartman H (2008) The Origin and Evolution of the Ribosome. Biol Direct 3:16.
26. Wolf YI, Koonin EV (2007) On the Origin of the Translation System and the Genetic Code in the RNA World by Means of Natural Selection, Exaptation, and Subfunctionalization. Biol Direct 2:14.
27. Woese CR (2001) Translation: In Retrospect and Prospect. RNA 7:1055-1067.
28. Fox GE (2010) Origin and Evolution of the Ribosome. Cold Spring Harb Perspect Biol 2:a003483.
29. Hury J, Nagaswamy U, Larios-Sanz M, Fox GE (2006) Ribosome Origins: The Relative Age of 23S rRNA Domains. Orig Life Evol Biosph 36:421-429.
30. Sloof P, Van den Burg J, Voogd A, Benne R, Agostinelli M, Borst P, Gutell R, Noller H (1985) Further Characterization of the Extremely Small Mitochondrial Ribosomal RNAs from Trypanosomes: A Detailed Comparison of the 9S and 12S RNAs from Crithidia Fasciculata and Trypanosoma Brucei with rRNAs from Other Organisms. Nucleic Acids Res 13:4171-4190.
A) Secondary structure of the 23S rRNA of the large subunit of T. thermophilus adapted with permission from Harry Noller. The six secondary structural domains of 23S rRNA are shown: Domain I in gray, Domain II in brown, Domain III in pink, Domain IV in yellow, Domain V in purple and Domain VI in orange. B) Tertiary interactions (dark blue) and phosphate-magnesium-phosphate linkages within Domain III. Each first shell magnesium-phosphate interaction is indicated by a magenta circle. The lines between the circles are the phosphate-magnesium-phosphate linkages. C) SHAPE reactivities for Domain IIIalone in 250 mM Na+. The blue nucleotides are unreactive. D) Magnesium-dependent SHAPE reactivities for Domain IIIalone, observed upon addition of 10 mM Mg2+. Only the nucleotides with the greatest proportional change in reactivity are indicated.
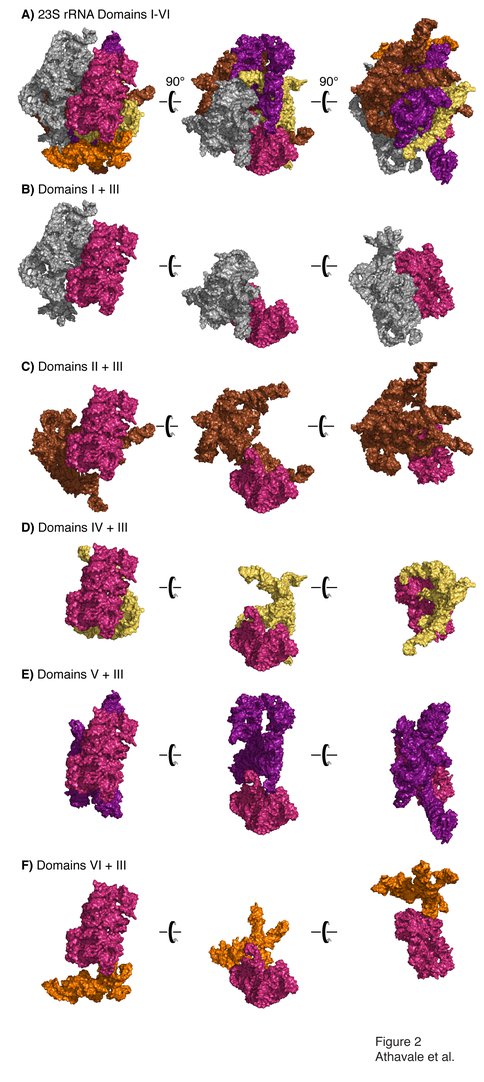
Domain III is compact and is not penetrated by other 2° domains. A) All six 2° domains of the 23S rRNA are shown, colored as in Figure 1. Three views, with a relative rotation of 90°, are shown. Panels B-F show interactions of Domain III with Domains I, II, IV, V and VI, respectively.
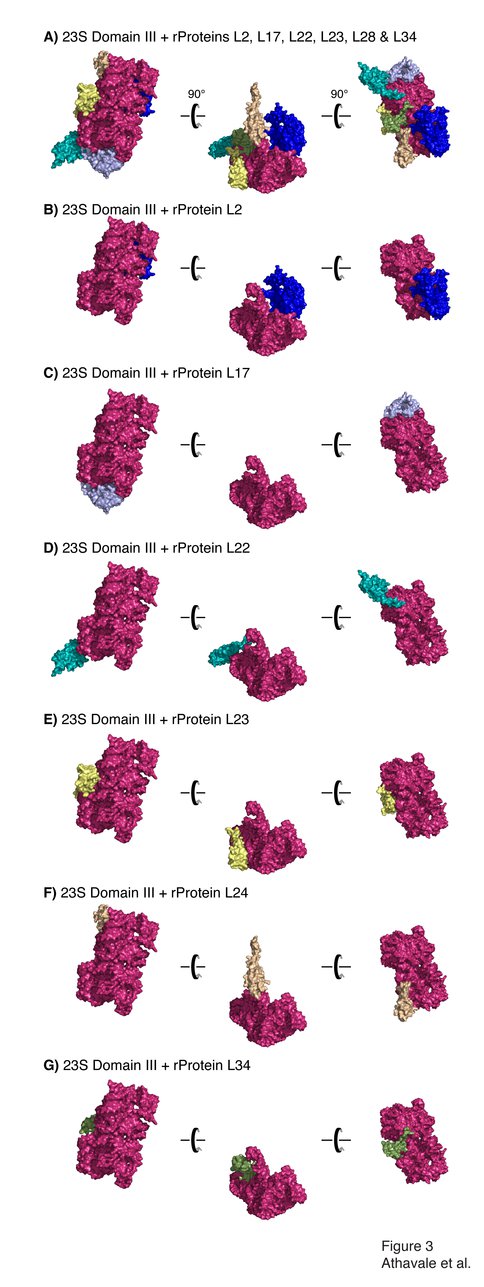
Domain III is not penetrated by ribosomal proteins. A) Domain III, colored and oriented as in Figure 2, with rProteins L2 (dark blue), L17 (light blue), L22 (dark green), L23 (yellow), L24 (light brown) and L34 (light green). Panels B-G show interactions of Domain III with each of these rProteins.
Figure 4. SHAPE reactivity for Domain IIIalone (blue) and Domain III23S (red). The vertical axis represents SHAPE reactivities and the horizontal axis represents nucleotide position using conventional E. coli numbering scheme. A) Domain IIIalone and Domain III23S in 250 mM Na+. B) Domain IIIalone and Domain III23S in 250 mM Na+ and 10 mM Mg2+. The inter-domain interactions between Domain III and Domains I, II and IV that cause differences in SHAPE reactivity between Domain IIIalone and Domain III23S are highlighted. Hydrogen bonds are shown by dashed lines, stacking interactions are shown by hashing and van der Waals contacts are shown by broad shaded arrows.
Publications
- Athavale, S.S., Gossett, J.J., Hsiao, C., Bowman, J.C., O’Neill, E., Hershkovitz, E., Preeprem, T., Hud, N.V., Wartell, R.M., Harvey, S.C. & Williams, L.D. (2012). Domain III of the T. Thermophilus 23S rRNA Folds Independently to a near-Native State. RNA.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Shreyas Athavale
Postdoc
Eli Hershkovits
Postdoc
Chiaolong Hsiao
Postdoc
Jessica Bowman
Research Staff
Eric O'Neill
Research Staff
Jared Gossett
Graduate Student
Thanawadee Preeprem
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.2
Production of complex life.