2009 Annual Science Report
 Pennsylvania State University
Reporting | JUL 2008 – AUG 2009
Pennsylvania State University
Reporting | JUL 2008 – AUG 2009
Developing New Biosignatures
Project Summary
The development and experimental testing of potential indicators of life is essential for providing a critical scientific basis for the exploration of life in the cosmos. In microbial cultures, potential new biosignatures can be found among isotopic ratios, elemental compositions, and chemical changes to the growth media. Additionally, life can be detected and investigated in natural systems by directing cutting-edge instrumentation towards the investigation of microbial cells, microbial fossils, and microbial geochemical products. Over the next five years, we will combine our geomicrobiological expertise and on-going field-based environmental investigations with a new generation of instruments capable of revealing diagnostic biosignatures. Our efforts will focus on creating innovative approaches for the analyses of cells and other organic material, finding ways in which metal abundances and isotope systems reflect life, and developing creative approaches for using environmental DNA to study present and past life.
Project Progress
Subproject: Raman imaging of kerogen and microfossils as a tool for Astrobiology
The Schopf laboratory pushed forward on developing Raman studies of preserved organic material. Results of detailed organic geochemical analyses of kerogens comprising Eocene (~45-Ma-old) plant axes have been published (2009, Organic Geochemistry 40: 353-364). Laboratory studies of the thermal alteration of ancient (Precambrian) kerogens are ongoing, in collaboration with Yongchun Tang at CalTech.
Subproject: Inorganic chemical biosignatures in Mars-analogue soils
Investigation of microbial communities on Svalbard, a Mars analogue (current work being done by Laura Liermann, research assistant)
Libby Hausrath, a former PSU graduate student, has investigated basalt and olivine weathering in a Mars analog environment, the Sverrefjell volcano in Svalbard (Hausrath et al., 2008). Weathered basalt samples were collected for analysis of biotic and abiotic weathering, and the potential identification of biomarkers. In addition, olivine and glass samples were buried at selected sites on the volcano (Figure 1). XPS and SEM analyses of the retrieved samples showed surface alterations, which were interpreted as indicating biological activity. Recently we have successfully extracted DNA from samples collected on Sverrefjell, including sites where olivine and glass samples were buried. We are currently doing PCR on the DNA samples, with the goal of generating clone libraries and sequencing to determine microbial community composition at these sites, and the potential impact of microorganisms on mineral weathering in this environment.

Figure 1. Sverefjell volcano, on Svalbard, a Mars analogue site. Letters indicate
where samples were collected, and numbers indicate where samples of forsterite
(olivine) and glass were buried, then retrieved one year later.
Studies of granite and basalt weathering with and without organic ligands (current work by Maya Bhatt, postdoctoral scholar, and Laura Liermann, research assistant)
Plants and soil microorganisms produce organic acids, which can affect the mobility of elements, and thus mineral weathering rates, in the soil environment. This leaching of elements by organic acids may produce trace element biosignatures in the rock record. To investigate this possibility, long-term (> 5 years) column experiments with powdered granite and basalt, with and without 0.01 M citrate at pH 6, are ongoing. Results from the first 45 weeks show that element dissolution rates from both granite and basalt are increased in the presence of citrate compared to controls without the ligand, and that the enhanced release of individual elements correlated with the stability constant of the element-citrate complex (Hausrath et al., 2009). Furthermore, elements whose dissolution from the minerals were enhanced by citrate have been reported to be enriched in organic-rich rivers, suggesting a leaching-redeposition pathway that may have left elemental biosignatures in the rock record. Analysis of additional column solution samples indicate that dissolution has reached steady-state. Samples of powders will be removed from the columns for chemical and mineralogical analyses and comparisons made between citrate and control columns to assess organomarkers of weathering. In addition, isotopic signatures of multiple elements (e.g. Fe, Cu, Mo) will be measured to determine the potential for isotopic biosignatures in this system.
Investigation of jarosite as a biosignature (work by Bryn Kimball, graduate student)
Detection of biosignatures improves our characterization of the existence of life under varying conditions. Extremophilic microorganisms, living under conditions such as extremely high or low temperatures, high pressures, high salinity, or low pH and high metal concentrations, are believed to exhibit ancient metabolisms, which were required for the relatively harsh conditions of early earth (Lal, 2008). Our work relates to using the mineral jarosite as a biosignature of microbial Fe-oxidation. We grew Acidithiobacillus ferrooxidans, an Fe- and S-oxidizing Proteobacterium, in an FeSO4-based medium at pH 2 and 22 ± 2°C. Over the course of 8 days, precipitates that formed in the presence of the bacteria became increasingly crystalline based on micro-X-ray diffraction analysis. These precipitates were jarosite. In comparison, in parallel abiotic experiments, precipitates that formed were amorphous. We conclude that under the same conditions, biogenic jarosite forms at a faster rate than abiogenic jarosite; thus, in systems that are open with respect to fluid flow, jarosite may be a biosignature. Further work is needed to characterize any physical and chemical differences between abiogenic and biogenic jarosite before this mineral can be used as a biosignature of microbial Fe-oxidation. This work is included in a manuscript that is in preparation.
Subproject: Using Methanosarcina to develop new biosignatures
Dr. Greg Ferry is leading efforts aimed at developing new biosignatures based on Methanosarcina as a model organism. These efforts include microbial polyphosphate as a biosignature and microbial methyl-sulfides as biosignatures.
Long chain poly-P is ubiquitous to microbial life on Earth (Proc. Natl. Acad. Sci. USA (2004) 101:16085), although few investigations have been reported in species from the Archaea domain. A role for poly-P in stress response has been suggested for the methane-producing archaeon Methanosarcina acetivorans (J. Proteome Res. (2007) 6:759). The probable ancient origin and widespread occurrence in diverse microbes, coupled with the role in coping with extreme environments, identifies poly-P as a facile biosignature for exploration of extraterrestrial life. We are developing poly-P for use as a biosignature by investigating poly-P metabolism in the Archaea domain with M. acetivorans for which the genome is annotated with genes encoding two putative poly-P kinases (Ppk1 and Ppk2) catalyzing synthesis of poly-P. One of the tasks identified is determination of factors controlling the accumulation of poly-P. This was investigated by determining the levels of long and short chain poly-P when M. acetivorans is grown with different substrates (please see the accompanying table). The results indicate that growth on methylotrophic substrates leads to greater production of long-chain poly-P versus growth with acetate. Currently, electrophoretic analysis is being used to determine the different chain lengths of short and long poly-P.
Growth substrate |
Long chain Poly-P |
Short chain Poly-P |
trimethylamine |
10.8 +1.5 |
ND |
methanol |
8.7 +1.5 |
0.148 + 0.02 |
acetate |
0.612 +.12 |
0.171 +0.02 |
ND, below limit of detection |
||
In order to determine the role of poly-P, mutants of M. acetivorans defective in synthesis of the poly-P kinases are under construction for evaluation of phenotypes indicative of function. Thus far, the ppk2 gene and flanking sequences were PCR amplified and ligated into the pJK301 vector that will be used to transform M. acetivorans and, by homologous recombination, disrupt the gene. Progress has also been made on construction of strains for which the poly-P kinase genes are under control of a tetracycline sensitive promoter that will allow controlled expression to further evaluate the physiological functions. Plasmids and M. acetivorans strains needed for the regulated expression were obtained from Dr. W. W. Metcalf. Both genes and the upstream sequences were PCR amplified and cloned into the pGK050A vector for eventual transformation into M. acetivorans.
To further explore the role of the two poly-P kinases, the enzymes will be over-produced in Escherichia coli and characterized. To this end, we have amplified both genes by PCR and cloned them into the E. coli expression vector pET22b.
The production of methyl sulfide during growth of methanogens with CO (Appl. Environ. Microbiol. (2008) 74: 540) and a role for methyl sulfide during anaerobic methane oxidation (Environ. Microbiol. (2008) 10: 162) has been postulated to be of biogeochemical importance with potential for development of new biosignatures. The enzymology of Methanosarcina acetivorans is being investigated to better understand the fundamental physiology of the metabolism of methyl sulfides by methanogens; specifically, enzymes proposed in the literature to catalyze the production and/or utilization of the methyl sulfides. One of three homologous genes (MA4558) from M. acetivorans postulated to react with methyl sulfides based on genetic analyses (Arch. Microbiol. (2007) 188: 463; Mol Microbiol (2009) 74: 227) was cloned and the protein produced in Escherichia coli for the purpose of validating the suspected activity and evaluating the kinetic parameters. The purified apoprotein lacking the postulated corrinoid cofactor was reconstituted with methylcobalmin. The UV-visible spectrum before and after reconstitution clearly indicates incorporation of the cofactor, and metal analyses indicate a cobalt to protein molar ratio of 1:1 indicating the presence of 1 corrinoid factor bound to each apoprotein molecule. Small amounts of the reconstituted protein were produced; thus, methods were developed to obtain milligram amounts for characterization. The apoprotein is currently being reconstituted with methylcorrinoid with the goal of testing for the suspected activity.
Subproject: Organic material and its associated isotopic signature
Graduate student Karen Whelley initiated lab work on abiotic synthesis and also worked with the Goddard team for the summer on organic material in meteorites. We also published several papers outlining advances in the application of secondary ion mass spectrometry on microorganisms (House et al., 2009; Orphan et al., 2009; Orphan and House, 2009).
Subproject: Isotopic biosignatures in minerals
The generation and recording of isotopic biosignatures in minerals is the general focus of this subproject. We are specifically interested in determining if microbes interact with their surrounding environment in a manner that affects the isotopic composition of minerals. To begin this investigation, we conducted field sampling in June 2009 of gypsum (as well as host limestone) found in close association with microbes in a sulfidic, carbonate-hosted cave (Grotta Bella) in the Frasassi cave system, Italy. Filtered and acidified water samples were also collected during this time, from springs reflecting deep sulfidic groundwater (Fissure Spring), springs affected by biogeochemical processes within the caves (multiple), and stream water integrating the larger watershed (Santino River). The calcium isotopic compositions of these samples were measured over a two-week period by Fantle at IFM-GEOMAR in Kiel, Germany in July 2009. This work marked the beginning of a new collaboration between Prof. Anton Eisenhauer and Fantle. The sulfur isotope compositions of the gypsum minerals were measured in September 2009 by Yumiko Watanabe at Penn State University (Ohmoto laboratory).
A map of the study area is shown in Figure 2, while images of the cave site and stream sampling protocol are shown in Figure 3. The interpretation of the isotope data (not shown) is preliminary and awaits additional analyses and geochemical modeling (on-going). It is clear from other studies of carbonate-hosted sulfidic caves that there is an intimate association between so-called “snottite” microbial communities and the gypsum that precipitates [Hose et al, 2000, Chem Geol 169: 399-423]. Such a relationship, though not imaged specifically at Grotta Bella during this field campaign, is suggested by the Ca and S isotope data. Future work, both experimental and theoretical, will examine the hypothesis that microbes fractionate Ca-S isotopes uniquely compared to abiotic processes.
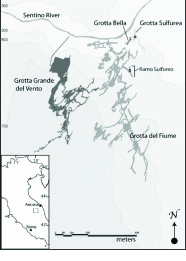
Figure 2. Map of the Frasassi cave system, Italy [Macalady et al., 2006]. Sampling was conducted in June 2009 in the Grotta Bella cave and the Sentino River, noted on the map.
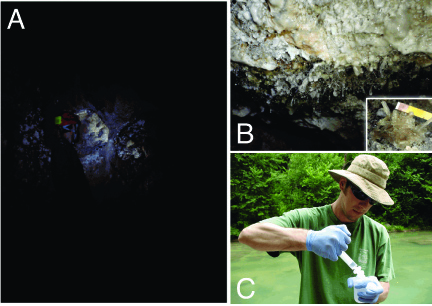
Figure 3 – Images of field sampling in Grotta Bella cave (June 2009). [A] Fantle at Owl Pit, from which gypsum samples associated with snottite biofilms were collected [B]. [C] Fantle collecting filtered spring and stream samples from outside the cave.
Subproject: DNA as a biosignature
Dr. Beth Shapiro, Dr. Duane Froese (U of Alberta), Michael Avery (PhD candidate). Drs Shapiro and Froese and Mr Avery spent several weeks identifying appropriate field locations in the Klondike gold fields of the northern Yukon Territory, Canada (Figure 4, 5 and 6). The team identified four potential drill locations, and have secured permission to drill at these location during next year’s field season. In the laboratory, Dr. Shapiro has been working to refine methods to separate potentially inhibitory compounds from soil samples and to design an appropriate test for radiation-related decay. Dr. Shapiro was also part of a team to develop a statistical model for age-based DNA decay that can be used to estimate the amount and rate of DNA decay occurring in a population sample through time (Rambaut A, Drummond AJ, Ho SYW, and Shapiro B. Accommodating the effect of ancient DNA damage on inferences of demographic histories. Mol. Biol. Evol. 26: 245-248, 2009), and to investigate the use of amino acid racemization as a method to predict DNA survival in ancient materials (Collins MJ, Penkman KEH, Rohalnd N, Shapiro B, Dobberstein RC, Rotz-Timme A, Hofretier M. Is amino acid racemization a useful tool for screening for ancient DNA in bone? Proc Roy Soc B 276: 2971-2977, 2009).

Figure 4. Field team inspects Pleistocene sediments in the Klondike gold fields of
the northern Yukon Territory, Canada.

Figure 5. Further scouting for appropriate field sites in the Klondike gold fields of
the northern Yukon Territory, Canada.

Figure 6. Michael Avery (PSU Ph.D. candidate) tries the massive hose at the Klondike gold fields of the northern Yukon Territory, Canada.
The House lab is working to develop DNA as a biosignature by focusing on DNA buried deep in marine sediment. Graduate student Mandi Martino has been working on developing a new method for amplifying DNA from very low quantities in a way that is suitable for metagenomics. This work is progressing well. She is also continuing to extract DNA from various sites along the Equatorial Pacific from the expedition with which she was involved (Figure 7).

Figure 7. Penn State Graduate student Mandi Martino out to sea in the Equatorial Pacific.
Publications
-
Cameron, V., Vance, D., Archer, C., & House, C. H. (2009). A biomarker based on the stable isotopes of nickel. Proceedings of the National Academy of Sciences, 106(27), 10944–10948. doi:10.1073/pnas.0900726106
-
Collins, M. J., Penkman, K. E. H., Rohland, N., Shapiro, B., Dobberstein, R. C., Ritz-Timme, S., & Hofreiter, M. (2009). Is amino acid racemization a useful tool for screening for ancient DNA in bone?. Proceedings of the Royal Society B: Biological Sciences, 276(1669), 2971–2977. doi:10.1098/rspb.2009.0563
-
Czaja, A. D., Kudryavtsev, A. B., Cody, G. D., & William Schopf, J. (2009). Characterization of permineralized kerogen from an Eocene fossil fern. Organic Geochemistry, 40(3), 353–364. doi:10.1016/j.orggeochem.2008.12.002
-
Hausrath, E. M., Neaman, A., & Brantley, S. L. (2009). Elemental release rates from dissolving basalt and granite with and without organic ligands. American Journal of Science, 309(8), 633–660. doi:10.2475/08.2009.01
-
Hausrath, E. M., Treiman, A. H., Vicenzi, E., Bish, D. L., Blake, D., Sarrazin, P., … Brantley, S. L. (2008). Short- and Long-Term Olivine Weathering in Svalbard: Implications for Mars. Astrobiology, 8(6), 1079–1092. doi:10.1089/ast.2007.0195
-
House, C. H. (2009). The Tree of Life Viewed Through the Contents of Genomes. Methods in Molecular Biology, None, 141–161. doi:10.1007/978-1-60327-853-9_8
-
House, C. H., Orphan, V. J., Turk, K. A., Thomas, B., Pernthaler, A., Vrentas, J. M., & Joye, S. B. (2009). Extensive carbon isotopic heterogeneity among methane seep microbiota. Environmental Microbiology, 11(9), 2207–2215. doi:10.1111/j.1462-2920.2009.01934.x
-
Orphan, V. J., & House, C. H. (2009). Geobiological investigations using secondary ion mass spectrometry: microanalysis of extant and paleo-microbial processes. Geobiology, 7(3), 360–372. doi:10.1111/j.1472-4669.2009.00201.x
-
Orphan, V. J., Turk, K. A., Green, A. M., & House, C. H. (2009). Patterns of 15 N assimilation and growth of methanotrophic ANME-2 archaea and sulfate-reducing bacteria within structured syntrophic consortia revealed by FISH-SIMS. Environmental Microbiology, 11(7), 1777–1791. doi:10.1111/j.1462-2920.2009.01903.x
-
Rambaut, A., Ho, S. Y. W., Drummond, A. J., & Shapiro, B. (2008). Accommodating the Effect of Ancient DNA Damage on Inferences of Demographic Histories. Molecular Biology and Evolution, 26(2), 245–248. doi:10.1093/molbev/msn256
-
Schopf, J. W. (2009). The hunt for Precambrian fossils: An abbreviated genealogy of the science. Precambrian Research, 173(1-4), 4–9. doi:10.1016/j.precamres.2009.01.003
-
Schopf, J. W., & Bottjer, D. J. (2009). World summit on ancient microscopic fossils. Precambrian Research, 173(1-4), 1–3. doi:10.1016/j.precamres.2009.06.001
-
Schopf, J. W., & Kudryavtsev, A. B. (2009). Confocal laser scanning microscopy and Raman imagery of ancient microscopic fossils. Precambrian Research, 173(1-4), 39–49. doi:10.1016/j.precamres.2009.02.007
-
Wander, M. C. F., & Schoonen, M. A. A. (2008). Reduction of N2 by Fe2+ via Homogeneous and Heterogeneous Reactions. Orig Life Evol Biosph, 38(2), 127–137. doi:10.1007/s11084-008-9122-9
-
Wander, M. C. F., Kubicki, J. D., & Schoonen, M. A. A. (2008). Reduction of N2 by Fe2+ via Homogeneous and Heterogeneous Reactions Part 2: The Role of Metal Binding in Activating N2 for Reduction; a Requirement for Both Pre-biotic and Biological Mechanisms. Orig Life Evol Biosph, 38(3), 195–209. doi:10.1007/s11084-008-9133-6
- Kimball, B. (2009). Biogeochemical Cycling of Copper in Acid Mine Drainage. Geosciences. University Park: The Pennsylvania State University.
- Lal, A.K. (2008). Origin of life. Astrophysics and Space Sceince, 317: 267-278.
- Schopf, J.W. & Bottjer, D.J. (2009). World Summit on Ancient Microscopic Fossils, Special Issue. Vol. 173. Precambrian Research.
- Schopf, J.W. (2009). Emergence of Precambrian paleobiology: a new field of science. In: Sepkoski, D. & Ruse, M. (Eds.). The Paleobiological Revolution, Essays on the Growth of Modern Paleontology. Chicago: University of Chicago Press.
- Schopf, J.W. (2009). Organismal evolution and radiation before the Cambrian. In: Ruse, M., Travis, J. & Zinser, J. (Eds.). Evolution: The First 4 Billion Years. Cambridge: Harvard University Press.
- Schopf, J.W. (2009). Paleontology, Microbial. In: Lederberg, J. & Schaechter, M. (Eds.). Encyclopedia of Microbiology, 3rd Edition. Amsterdam: Elsevier.
- Zerkle, A.L., Junium, C.K., Canfield, D.E. & House, C.H. (2008). Production of 15 N-depleted biomass during cyanobacterial N 2 -fixation at high Fe concentrations. J. Geophys. Res, 113(G03024).
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
George Cody
Collaborator
Andrew Czaja
Collaborator
Jason Dworkin
Collaborator
Duane Froese
Collaborator
Libby Hausrath
Collaborator
Yongchun Tang
Collaborator
Maya Bhatt
Postdoc
Suharti Suharti
Postdoc
Venkata Vepachedu
Postdoc
John Cantolina
Research Staff
Anatoliy Kudryavtsev
Research Staff
Laura Liermann
Research Staff
Dennis Walizer
Research Staff
Yumiko Watanabe
Research Staff
Zhidan Zhang
Research Staff
Michael Avery
Graduate Student
Bryn Kimball
Graduate Student
Lexan Lhu
Graduate Student
Mandi Martino
Graduate Student
Karen Smith
Graduate Student
-
RELATED OBJECTIVES:
Objective 2.1
Mars exploration.
Objective 2.2
Outer Solar System exploration
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems








