2009 Annual Science Report
 Pennsylvania State University
Reporting | JUL 2008 – AUG 2009
Pennsylvania State University
Reporting | JUL 2008 – AUG 2009
Biosignatures in Relevant Microbial Ecosystems
Project Summary
In this project, PSARC team members explore the isotope ratios, gene sequences, minerals, organic biomarkers, and other biosignatures in modern ecosystems that function as analogs for early earth ecosystems, or for life that may be present elsewhere in the solar system and beyond. Many of these environments are “extreme” by human standards and/or have conditions that are at the limit for microbial life on Earth.
Project Progress
Molecular signatures of microbial life in the Dead Sea
House, Freeman, & graduate student Rhodes (in collaboration with A. Oren & O. Beja)
Moshe Rhodes has traveled to the Dead Sea twice. He has worked on extensive DNA sequencing of the modern (2007) Dead Sea as well as the Dead Sea from a 1992 bloom event. Moshe has also developed computer programs to analyze the large amount of data we have collected. The work has generated a first paper that is now under review after an initial set of revisions. The paper is at Environmental Microbiology. The paper deals with amino acid bias of hypersaline genomes. It also uses sequence analysis coupled with amino acid bias to investigate which microbial groups are indigenous to the Dead Sea and what microorganisms have transferred genetic material to Dead Sea microbiota. We have a second paper in preparation, as well.
Impact cratering and subsurface microbial life
Macalady, Freeman, Shapiro & graduate student Kristine Korzow Richter (in collaboration with C. Cockell, D. Vanko & M. Voytek)
A new Ph.D. student (Fall 2009), Kristine Korzow Richter, will be co-advised by Beth Shapiro (primary) and Jenn Macalady to work on the metagenomics of authenticated core samples from the ~2 km deep Chesapeake Bay impact structure. A research meeting with USGS collaborators including Mary Voytek and Greg Gohn (head of the USGS/ICDP drilling project) has been set for Nov. 30 at the USGS. We plan to view the core lithologies and mineralogy and retrieve preserved microbiology/genomics samples to begin the project. Kristine is currently taking classes that will set the stage for her project and fulfill requirements of the Astrobiology Dual Ph.D. Degree program, as well as investigating methods of DNA extraction for rock materials in the Shapiro ancient DNA laboratory on campus.
Methane-derived isotopic & mineralogical biosignatures in cold seeps
Orphan, Ferry, Freeman, and McKeegan (in collaboration with J. Grotzinger)
Additional team members (Orphan lab): Jake V. Bailey (postdoc), Timothy D. Raub (postdoc), A. Nele Meckler (postdoc), Theresa M.D. Raub (postdoc), Benjamin K. Harrison (graduate student), Abigail M. Green (graduate student)
Preservation potential of microbial consortia in recent authigenic carbonates
Marine methane seeps are commonly associated with authigenic minerals such as carbonates and sulfides. The precipitation of these minerals is thought to be mediated by the anaerobic oxidation of methane – a process carried out by consortia of methanotrophic archaea and sulfate-reducing bacteria. The authigenic carbonates that result from AOM precipitate in porewaters with high concentrations of morphologically-distinctive cells and aggregates, making seep carbonates attractive targets for biosignature research – particularly the study of microbial cell taphonomy.
With the goal of determining whether methane seep endemic microbes are preserved as microfossils in authigenic seep precipitates, we undertook a survey of Quaternary authigenic carbonates from the Eel River Basin. Petrographic examination of carbonates revealed structures that bear close morphological resemblance to the bacterial and archaeal cell aggregates thought to mediate seep carbonate precipitation in surrounding sediments (Microstructure figured at right). However, additional analyses using SEM, EDS, Raman spectroscopy, and rock magnetic analyses suggest that these structures are not the fossilized remains of microorganisms at all, but rather the products of the diagenetic alteration of sulfide framboids. These results are important because the ability to differentiate between cellular remains and acellular mineral matter is critical for life detection efforts on other planets, as well as for tracking the evolution of biogeochemcial cycles on Earth.
In addition to the analytical approaches described above, the distribution of amorphous iron sulfide minerals entombed within the carbonates alongside with 12C, 13C, and CN- were mapped at a sub-micron scale using a nanoSIMS in hopes of better understanding their petrogenesis. However, efforts to use the nanoSIMS to measure the carbon isotope composition of organic matter associated with the microstructures was hampered by the presence of mounting epoxy injected into interstices during sample preparation of the porous carbonates. Future analyses will be conducted after sample preparation with C-free epoxy.
The results of the investigation of the seep-associated microstructures are detailed in a 43-page manuscript entitled “Pseudofossils in relict methane seep carbonates resemble endemic microbial consortia” by Bailey et al. This manuscript is currently in review at the journal Palaeogeography, Palaeoclimatology, Palaeoecology.
Life in Greenland glacial ice
Brenchley, Miteva, and Loveland-Curtze
We are developing and testing methods for direct quantitative analyses of the abundance, viability and diversity of Greenland ice core samples with variable cell biomass. We are using novel cultivation strategies to recover isolates and have characterized and described two new bacterial species. We are also performing comparative analyses of the microbial diversity at different depth of the Greenland ice sheet in relationship to climate.
Direct analysis of ice core samples
We continued to optimize our flow cytometry (FC) protocols for quantitative estimation of total cell abundance (SYTO13 staining), viability (LIVE/DEAD kit) and relative distribution of specific bacterial or archaeal microorganisms (FISH-FLOW (Fig. 1A and 1B). These studies are needed to minimize some limitations with flow cytometry such as the inability to separate strictly live from dead cells due to transitional stages and obtaining atypical results with environmental samples. Furthermore, the Penn State flow cytometry facility recently equipped its triple-laser Cytopeia Influx sorter with a small particle (<300 nm) detector that we started using to analyze and aseptically separate ultrasmall cells or other cell clusters (Fig.1C). Parameters were set with beads and cells of known sizes.
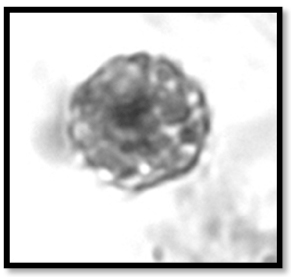
Fig1.. Examples of FC analyses: A. Cells with red fluorescence after hybridization with Cy5 labeled Crenarchaeota specific probes; B. Cells with strong green fluorescence after hybridization with FITC labeled eubacterial probe; C. Clusters of ultrasmall cells (arrow) detected on the Cytopeia Influx sorter in an aerobic enrichment for Crenarchaeota that may be separated individually.
Fig. 1. Examples of FC analyses: A. Cells with red fluorescence after hybridization with Cy5 labeled Crenarchaeota specific probes; B. Cells with strong green fluorescence after hybridization with FITC labeled eubacterial probe; C. Clusters of ultrasmall cells (arrow) detected on the Cytopeia Influx sorter in an aerobic enrichment for Crenarchaeota that may be separated individually.
These direct quantitative analyses provide rapid and sensitive cell-specific information for particular sizes and types of microorganisms within the microbial populations in our ice core samples and enrichments.
Description of two novel bacterial species isolated from Greenland ice
We characterized and described two ultramicrobacterial isolates from a 120,000-year-old, 3042 m deep Greenland glacier ice core as novel species in two papers:
Loveland-Curtze J., Miteva V., Brenchley J. (2009) Herminiimonas glaciei sp. nov., a novel ultramicrobacterium from 3042 m deep Greenland glacial ice. Intern. J. Syst. Evol. Microbiol. 59: 1272 – 1277. *This paper was a subject of a press release by the Society for General Microbiology (June 12, 2009) http://www.sgm.ac.uk/news/Releases and was covered in the May and August 2009 issues of Microbiology Today http://www.sgm.ac.uk/pubs/micro_today/pdf/050910.pdf), http://www.sgm.ac.uk/pubs/micro_today/pdf/080906.pdf and in the NSF news http://nsf.gov/news/news_summ.jsp?cntn_id=114995. .
Loveland-Curtze J., Miteva V., Brenchley J. (2009) Novel ultramicrobacterial isolates from a deep Greenland ice core represent a proposed new species, Chryseobacterium greenlandensis sp. nov. Extremophiles (submitted).
These are the only validly described species from Greenland ice and among the few from glacial ice.
Fig. 2. A. Chryseobacterium greenlandense; B. Herminiimonas glaciei.
Fig 2. A. Chryseobacterium greenlandense; B. Herminiimonas glaciei.
Relationship of microbial diversity at different depths to deposition climate Our findings were published in the Special issue on Polar microbiology of the journal Environmental Microbiology: Miteva V., Teacher C., Sowers T., Brenchley J. (2009) Comparison of microbial diversity at different depths of the GISP2 Greenland ice core in relation to deposition climate. Environ. Microbiol. 11, 3, 640-656.
These results suggested that microbiological records in ice cores may provide exciting new information about past climates and underscored the importance of further searching for spatial and temporal diversity patterns that could lead to microbial markers as new tracers of specific past climates. We anticipate improving our understanding on how local and global climate processes may have shaped the composition of the microbial populations transported and deposited over Greenland.
Biosignatures of life in pelagosite
Macalady, Schopf (in collaboration with A. Montanari, D. Bice)
Pelagosite is a dark, shiny, pisolitic mineral that encrusts rocky coasts exposed to sea spray. X-ray diffraction and optical microscopy showed that pelagosite is aragonite with two to three micron-thick, alternating dark-light laminae. Work on samples collected in parallel with this study showed that pelagosite laminae faithfully record Holocene climate cycles, inluding sunspot, North Atlantic Oscillation (NAO), and ENSO (Bice & Montanari, in preparation). Pelagosite crusts along the Adriatic Sea coast range in age from modern to perhaps tens of thousands of years old based on neotectonic estimates. A role for biology in the precipitation of pelagosite crusts is suggested by its growth on limestone with strong chemical corrosion textures (Figure 3). Using specimens provided by J. Macalady (PennState NAI team), UCLA studied pelagosite thin sections using optical microscopy, scanning electron microscopy, and Raman spectroscopy. Analysis by Raman and CLSM show that the darker layers (Figure 4) are not kerogen. The particulate material producing the darkness is very fine-grained, perhaps the particles are tiny trapped air bubbles or, perhaps, the darkness is caused by diffraction of the pelagositic crystals. Together with morphological observations on the layering, these results show that pelagosite crusts are surf-precipitated deposits, not microbially produced “microstromatolites”. However, abundant, large-molecular-weight DNA can be extracted from pelagosite, but not from calcite at the surface of the immediately adjacent limestone, indicating that aragonite encrustation preserves cells and/or biomolecules preferentially compared to the naked host rock. Nucleic acid analyses of pelagosite crusts indicate that they are intimately associated with Xenococcus, a poorly known cyanobacterial genus.
Fig. 3. Pelagosite (dark crusts and spots) developed on corroding limestone in the sea spray zone of the Adriatic Sea.
{300}}
Biosignatures of life in Archaean-analog anoxic biofilms
Macalady & graduate students McCauley, Jones (incollaboration with A. Montanari, A. Zerkle)
A rare window into an anoxic subsurface aquifer is provided by meromictic lakes deep with the sulfidic Frasassi cave complex in central Italy. Cave divers exploring a remote underwater passage in the stratified lakes discovered unusual, rope-like microbial biofilms in the lower, anoxic water layer. Geochemical data suggest that little redox energy is available for life, consistent with low signal from domain-specific FISH probes. The carbon isotope signatures of the biofilm (-33‰) and DIC (-9‰) indicate in situ production by lithoautotrophs using RuBisCO. Ribosomal clone libraries constructed from the biofilm are dominated by archaea in the enigmatic Marine Benthic Group D (MBG-D/DHVE-1) along with diverse sulfate reducing bacteria. Most of the remaining clones affiliate with 11 major uncultivated or novel prokaryotic lineages. Diverse dsrAB gene sequences were retrieved from the biofilm, consistent with diverse Deltaproteobacterial 16S rDNA clones and geochemistry. Methane is detectable in the anoxic water although no 16S rDNA sequences associated with known methanogens or anaerobic methane oxidizers were retrieved. mcrA gene sequences retrieved from the biofilm are not related to cultivated methanogens or to known anaerobic methane oxidizers. Our data suggest that novel archaea and bacteria, including MBG-D archaea, are important in the dark, energy-limited cave biofilm. These microorganisms and their potentially novel metabolic strategies are relevant for understanding biogeochemistry and biosignatures of non-photosynthetic, energy-limited environments on the modern and ancient Earth and elsewhere in the solar system.
A successful expedition to recover a second biofilm sample was carried out in September 2009 by Macalady, Jones, Italian cave explorers, and American cave diver and National Geographic Explorer Kenneth Broad (Rosenstiel School of Marine and Atmospheric Sciences, University of Miami) (Figure 5). The expedition provided new geochemical data (Figure 6) that will allow us to calculate free energy available from the environment of the biofilms. Astrobiology Dual Title Ph.D. student Becky McCauley is currently preparing the biofilm samples for metagenomic sequencing that will shed new light on the metabolisms of uncultivated archaea in the biofilm.
Fig. 5. Italian cave explorer Sandro Mariani (right) and American cave diver Kenneth Broad (left) prepare to collect samples of biofilms and water from the anoxic layer of a stratified cave lake 600 m below the ground surface in central Italy.
Fig. 6. Italian cave scientist Fabio Baldoni (left) and PSARC Ph.D. student Daniel S. Jones preserving samples of anoxic cave lake water for geochemical analyses.
Publications
-
Battistuzzi, F. U., & Hedges, S. B. (2008). A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land. Molecular Biology and Evolution, 26(2), 335–343. doi:10.1093/molbev/msn247
-
Beal, E. J., House, C. H., & Orphan, V. J. (2009). Manganese- and Iron-Dependent Marine Methane Oxidation. Science, 325(5937), 184–187. doi:10.1126/science.1169984
-
Hedges, S. B., Marin, J., Suleski, M., Paymer, M., & Kumar, S. (2015). Tree of Life Reveals Clock-Like Speciation and Diversification. Molecular Biology and Evolution, 32(4), 835–845. doi:10.1093/molbev/msv037
-
Hoashi, M., Bevacqua, D. C., Otake, T., Watanabe, Y., Hickman, A. H., Utsunomiya, S., & Ohmoto, H. (2009). Primary haematite formation in an oxygenated sea 3.46 billion years ago. Nature Geosci, 2(4), 301–306. doi:10.1038/ngeo465
-
Jones, D. S., Tobler, D. J., Schaperdoth, I., Mainiero, M., & MacAlady, J. L. (2010). Community Structure of Subsurface Biofilms in the Thermal Sulfidic Caves of Acquasanta Terme, Italy. Applied and Environmental Microbiology, 76(17), 5902–5910. doi:10.1128/aem.00647-10
-
Loveland-Curtze, J., Miteva, V. I., & Brenchley, J. E. (2009). Herminiimonas glaciei sp. nov., a novel ultramicrobacterium from 3042 m deep Greenland glacial ice. INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, 59(6), 1272–1277. doi:10.1099/ijs.0.001685-0
-
Loveland-Curtze, J., Miteva, V., & Brenchley, J. (2009). Novel ultramicrobacterial isolates from a deep Greenland ice core represent a proposed new species, Chryseobacterium greenlandense sp. nov.. Extremophiles, 14(1), 61–69. doi:10.1007/s00792-009-0287-6
-
Luo, G., Kump, L. R., Wang, Y., Tong, J., Arthur, M. A., Yang, H., … Xie, S. (2010). Isotopic evidence for an anomalously low oceanic sulfate concentration following end-Permian mass extinction. Earth and Planetary Science Letters, 300(1-2), 101–111. doi:10.1016/j.epsl.2010.09.041
-
Lyons, J. R. (2009). Atmospherically-derived mass-independent sulfur isotope signatures, and incorporation into sediments. Chemical Geology, 267(3-4), 164–174. doi:10.1016/j.chemgeo.2009.03.027
-
Miteva, V., Teacher, C., Sowers, T., & Brenchley, J. (2009). Comparison of the microbial diversity at different depths of the GISP2 Greenland ice core in relationship to deposition climates. Environmental Microbiology, 11(3), 640–656. doi:10.1111/j.1462-2920.2008.01835.x
-
Rhodes, M. E., Fitz-Gibbon, S. T., Oren, A., & House, C. H. (2010). Amino acid signatures of salinity on an environmental scale with a focus on the Dead Sea. Environmental Microbiology, 12(9), 2613–2623. doi:10.1111/j.1462-2920.2010.02232.x
-
Young, S. A., Saltzman, M. R., Foland, K. A., Linder, J. S., & Kump, L. R. (2009). A major drop in seawater 87Sr/86Sr during the Middle Ordovician (Darriwilian): Links to volcanism and climate?. Geology, 37(10), 951–954. doi:10.1130/g30152a.1
- Battistuzzi, F.U. & Hedges, S.B. (2009). Archaebacteria. In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Battistuzzi, F.U. & Hedges, S.B. (2009). Eubacteria. In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University press.
- Beal, E.J. (2009). Geochemical Requirements of the Anaerobic Oxidation of Methane in the Eel River Basin. Geosciences. The Pennsylvannia State University.
- Bhattacharya, D., Yoon, H-S., Hedges, S.B. & Hackett, J.D. (2009). Eukaryotes (Eukaryota). In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Hedges, S.B. & Kumar, S. (2009). Timetree (www.timetree.org) [Online]. Website: www.timetree.org.
- Hedges, S.B. & Vidal, N. (2009). Lizards, snakes, and amphisbaenians (Squamata). In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Hedges, S.B. (2009). Life. In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Hedges, S.B. (2009). Vertebrates (Vertebrata). In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Heinicke, M.P. (2009). A molecular phylogenetic perspective on the evolutionary history of terraranan frogs, a vertebrate mega-radiation. Department of Biology. University Park: The Pennsylvania State University.
- Heinicke, M.P., Naylor, J.P. & Hedges, S.B. (2009). Cartilaginous fishes (Chondrichthyes). In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Heinicke, M.P., Sander, J.M. & Hedges, S.B. (2009). Lungfishes (Dipnoi). In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Hoashi, M., Watanabe, Y. & Ohmoto, H. (2009). Primary hematite formation in an oxygenated deep sea 3.46 billion years ago. Goldschmidt conference. Davos, Switzerland.
- Horodyskyj, L. (2009). Soil formation and terrestrial biosignatures in the Middle Cambrian [electronic resource]. Geosciences. University Park: The Pennsylvania State University.
- Johnson, I. (2009). The Earth’s oldest (~3.4 GA) paleosol at Trendall Ridge in the North Pole Dome region of the Eastern Pilbara Craton, Western Australia. Geosciences. University Park: The Pennsylvania State University.
- Johnson, I., Watanabe, Y., Stewart, B. & Ohmoto, H. (2009). Earth’s oldest (~3.4 Ga) lateritic paleosol in he Pilbara Craton, Western Australia. Goldschmidt conference. Davos, Switzerland.
- Jones, D.S., Schaperdoth, I. & MacAlady, J.L. (2010). Microbial communities from extremely acidic sulfidic cave biofilms. Environmental Microbiology, submitted.
- Kump, L.R., Condie, K.C. & Arthur, M.A. (2009). Co-evolutionary implications of alternative models of plate tectonics in Earth history. Geol. Soc. America Ann. Meeting. Portland, OR.
- Kump, L.R., Meyer, K.M., Ridgwell, A. & Payne, J.L. (2009). Biosphere response to volcanic CO 2 release: Siberian Traps and End-Permian Mass Extinction. Geol. Soc. America Ann. Meeting. Portland, OR.
- Meyer, K.M., Kump, L.R., MacAlady, J., Schaperdoth, I. & Freeman, K. (in review). Benthic production of a putative planktonic biomarker. Geobiology.
- Miteva, V. (2009). Microorganisms associated with glaciers. In: V. P. Singh, P.S.a.U.K.H. (Eds.). Encyclopedia of Snow, Ice and Glaciers. Springer (submitted).
- Ohmoto, H. (2009). Redox evolution of volcanic gas through geologic time. Goldschmidt conference. Davos, Switzerland.
- Ohmoto, H., Bevacqua, D.C. & Watanabe, Y. (2009). Alteration of submarine volcanic rocks in oxygenated Archean oceans. American Geophysical Union Annual Meeting. San Francisco, CA.
- Saltzman, M.R., Young, S.A., Kump, L.R., Foland, K.A. & Leslie, S. (2009). The Late Ordovician glaciation and mass extenction: Relation to basaltic weathering and volcanic degassing. Geol. Soc. America Ann. Meeting. Portland, OR.
- Urban, N., Bralower, T., Keller, K. & Kump, L.R. (2009). Statistical interpretation of the rate of carbon isotope changes at the onset of the Paleocene-Eocene Thermal Maximum. American Geophysical Union Fall Meeting. San Francisco, CA.
- Vidal, N., Rage, J-C., Coulous, A. & Hedges, S.B. (2009). Snakes (Serpentes). In: Hedges, S.B. & Kumar, S. (Eds.). The Timetree of Life. New York: Oxford University Press.
- Yamaguchi, K.E., Kato, Y., Nakamura, K., Suzuki, K., Watanabe, Y., Nedachi, M. & Ohmoto, H. (2009). REE+Y geochemistry of the 3.46 Ga Marble Bar Chert recovered by the Archean Biosphere Drilling Project. Goldschmidt Conference. Davos, Switzerland.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
James Ferry
Co-Investigator
Katherine Freeman
Co-Investigator
James Schopf
Co-Investigator
Beth Shapiro
Co-Investigator
Oded Beja
Collaborator
David Bice
Collaborator
Kenneth Broad
Collaborator
Charles Cockell
Collaborator
Sorel Fitz-Gibbon
Collaborator
Kevin McKeegan
Collaborator
Alessandro Montanari
Collaborator
Aharon Oren
Collaborator
John Spear
Collaborator
Mary Voytek
Collaborator
Aubrey Zerkle
Collaborator
Jake Bailey
Postdoc
Tim Raub
Postdoc
Theresa M. D. Raub
Postdoc
Jennifer Loveland-Curtze
Research Staff
Vanya Miteva
Research Staff
Irene Schaperdoth
Research Staff
Zhidan Zhang
Research Staff
Heidi Albrecht
Doctoral Student
Daniel Jones
Doctoral Student
Kristine Korzow Richter
Doctoral Student
Rebecca McCauley
Doctoral Student
Moshe (Mathew) Rhodes
Doctoral Student
Abigail M. Green
Graduate Student
Benjamin Harrison
Graduate Student
Norbert Lazar
Graduate Student
Thomas Roberts
Graduate Student
Labdhi Seth
Graduate Student
Anatoliy Kudryavtsev
Unspecified Role
A. Nele Meckler
Unspecified Role
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.3
Effects of extraterrestrial events upon the biosphere
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems