2009 Annual Science Report
 Massachusetts Institute of Technology
Reporting | JUL 2008 – AUG 2009
Massachusetts Institute of Technology
Reporting | JUL 2008 – AUG 2009
Origins of Multicellularity
Project Summary
One of the earliest events in the evolution of animals was the origin of multicellularity. By studying the closest living relatives of animals, the choanoflagellates, we can begin to reconstruct the biology of the unicellular ancestors from which all living animals evolved.
Project Progress
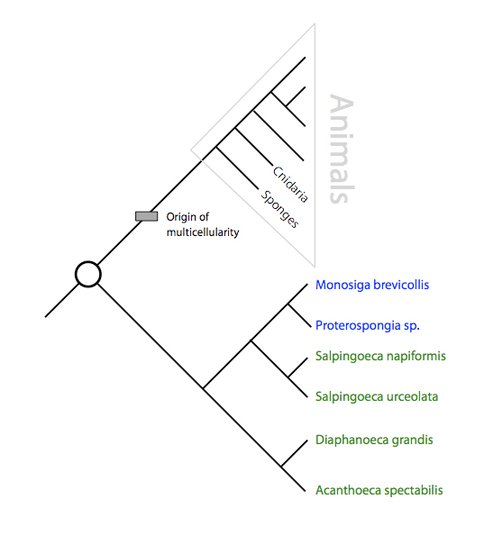
To gain insight into the origin of animal multicellularity, we aim to reconstruct the genome of the last common ancestor of animals and their sister group, the choanoflagellates. While genome sequences of diverse animals are available, only two choanoflagellates (Monosiga brevicollis and Proterospongia sp.) have been sequenced to date and they each show evidence of genome reduction and gene loss ([1]; www.broadinstitute.org/annotation/genome/multicellularity_project/Info.html). Therefore, we are preparing to sequence the genomes and transcriptomes of diverse choanoflagellates using new deep-sequencing technologies (e.g. Illumina sequencing).
There are three important technical challenges to sequencing choanoflagellate genomes using Illumina technology. The first challenge is to optimize culture conditions for each species to allow cultivation of large quantities of choanoflagellate cells. We have now done this for four species that represent choanoflagellate clades not previously sequenced: Salpingoeca napiformis, Salpingoeca urceolata, Diaphanoeca grandis, and Acanthoeca spectabilis [2]. The second challenge is to enrich for choanoflagellate DNA over that of their abundant co-cultured bacterial prey. Using a combination of antibiotic treatment and rapid culturing, we have reduced the bacterial load for each species in culture and improved the ratio of choanoflagellate:bacterial DNA. Finally, the ability to sequence and assemble a eukaryotic genome de novo using the short reads generated by Illumina technology is relatively unproven. Using short reads from the sequencing of a sponge genome, we have utilized emerging algorithms to assemble contigs of sufficient size to detect protein-coding genes (Nichols, Richter, and King, unpublished). We have therefore surmounted each of the major technical challenges associated with rapidly sequencing new choanoflagellate genomes. Sequencing, assembly, and annotation of four new choanoflagellate genomes is expected in the coming year.
Figure 1. Genomic ancestry of choanoflagellates and animals. Figure 1. Genomic ancestry of choanoflagellates and animals
By comparing the genomes of diverse choanoflagellates with each other and with those of animals, it is possible to reconstruct the genome of their common ancestor (circle). Choanoflagellates with sequenced genome (blue) and genome projects currently in progress (green).
As a complement to the global genome-sequencing approach, we are performing a more focused study on the ancestry of the hedgehog signaling pathway. The hedgehog protein, a key regulator of developmental patterning in bilaterian animals, signals through the primary cilium which is equivalent to the choanoflagellate flagellum [3,4]. We have identified core components of the hedgehog signaling pathway in M. brevicollis that were previously known only from animals [1,5,6]. Although a direct homolog of the hedgehog signaling ligand is absent in choanoflagellate, we have identified a membrane-integral cadherin protein (hedgling/MBCDH11) that has structural homology to the hedgehog signal domain [5]. We are currently generating antibodies against MBCDH11 in order to test two discrete hypotheses: 1) that the N-terminal region with homology to the hedgehog signaling ligand is cleaved and functions as a diffusible signal, and; 2) that the integral-membrane portion of the protein localizes to the apical flagellum — a pattern that would be consistent with an ancient signaling role for both the hedgling protein and the primary cilium. M. brevicollis provides a simple system in which to study the ancestral function of hedgehog signaling and promises to yield insight into the role of this important developmental signaling pathway during the transition to multicellularity.
Publications
-
Carr, M., Leadbeater, B. S. C., Hassan, R., Nelson, M., & Baldauf, S. L. (2008). Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proceedings of the National Academy of Sciences, 105(43), 16641–16646. doi:10.1073/pnas.0801667105
-
PROJECT INVESTIGATORS:
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.