2009 Annual Science Report
 Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Molecular Resurrection of the Ancestral Peptidyl Transferase Center
Project Summary
We have designed, and are resurrecting, a model of the a-PTC (ancestral Peptidyl Transferase Center), which we believe to be around 4 billion years old. The proposed a-PTC contains around 600 nucleotides of ancestral ribosomal RNA (a-rRNA), three ancestral ribosomal peptides (a-rPeptides), and inorganic cations, all of which are relatively straightforward to obtain or produce. The results of our molecular resurrection will allow one to test ideas about primitive living systems, including the origin of protein.
Project Progress
Our ultimate goal is to resurrect the ancestral Peptidyl Transferase Center (a-PTC), to create it in silico, in vitro and in vivo, and to explore its assembly, enzymatic activity and interactions with other extant and ancestral components. The resurrected a-PTC will allow us test and evaluate putative chronologies of ribosomal evolution and to recreate early steps in protein biosynthesis. Thus far we have developed two- and three-dimensional models of the a-PTC, which is composed primarily of a-rRNA (ancestral rRNA), but also contains three fragments of ancestral ribosomal proteins (a-rPeptides) and highly coordinated Mg2+ ions.
To develop a-PTC models, we have surveyed a wealth of published structural, biochemical, phylogenic and biological data that is too extensive to review here. We have analyzed molecular interactions1; RNA-RNA, RNA-protein and RNA-Mg2+ ion. We used high resolution three-dimensional LSU structures from bacteria2 and archaea.3 We performed exhaustive and recursive structural alignment and that gave an accurate unbiased superimposition4 of the LSU structures of H. marismortui and the T. thermophilus. In addition we used sequence comparisons of ribosomal proteins1 and of rRNA.5 We incorporated a new analysis of ribosomal evolution by Bokov, based on A-minor interactions.6 These orthogonal analytical methods reach strong consensus on the basic elements of the a-PTC. For example critical Mg2+-RNA interactions, identified by coordination geometry alone, involve ribosomal secondary elements that are absolutely conserved throughout the phylogenic tree, including in cytoplasmic, mitochondrial and chloroplast ribosomes from all living organisms
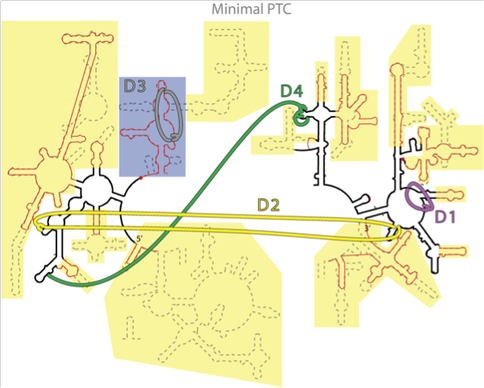
Figure 1. Figure 1. Consensus between phylogeny and Mg2+-RNA interactions. The putative secondary structure of a-rRNA (black line) along with the secondary structures of 23S rRNA (equivalent) of the mitochondrion of B. taurus (red line) and T. thermophilus (dashed line). Critical Mg2+ interactions are indicated by broad colored lines. The yellow and gray shaded areas indicate the secondary structural elements that have been cut off somewhere in the phylogenetic tree. The blue shaded area indicates the structure that we believe folds and assembles independently of the a-PTC, with an isolated Mg2+ cluster.
and are identified by A-minor analysis as aboriginal (not shown).
To resurrect the a-PTC in vitro we sliced off the 'non-ancestral’ regions of the 23S rRNA of T. thermophilus, and pasted the remaining RNA fragments together, forming one contiguous RNA molecule. This process was conducted at the DNA level. We constructed the a-rRNA gene using overlapping PCR, a technique for simultaneously linking and amplifying multiple (up to 20) oligonucleotides.
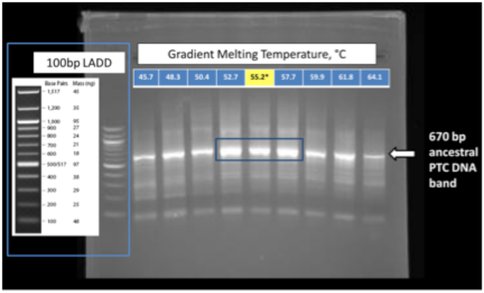
Figure 2. Figure 2. Diagnostic gel of PCR products. Shown is the 2% agarose gel following simultaneous synthesis and amplification of the DNA gene for the a-rRNA. The blue box indicates the full range of conditions, and the yellow box shows the optimal conditions. The arrow indicates the location of a 670 bp DNA marker.
The PCR product was cloned to pUC19 plasmid, sequenced, and used to transform DH5α cells. The a-rRNA was prepared by in vitro transcription.
Thus far we have successfully synthesized one native a-rRNA and two a-rRNA mutant transcripts.
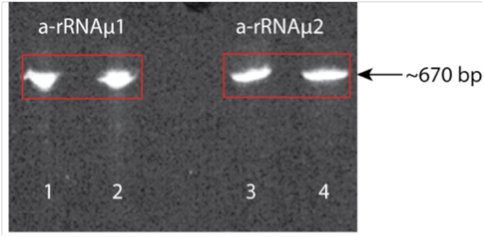
Figure 3. Figure 3. Shown are two a-rRNA mutants (~670 bp) on a 5% acrylamide, 8M urea gel. Lanes 1 and 2 are a-rRNA mutant 1 (a-rRNAµ1); lanes 3 and 4 are a-rRNA mutant 2 (a-rRNAµ2). In lanes 2 and 4 the a-rRNA was heated at 80 ºC for 3 min. and quick cooled. In lanes 1 and 3 the rRNA was loaded after purification without heating/quick cooling.
Our current goal is to (i) characterize the a-rRNA folding and assembly with a-rPeptides and Mg2+ ions (to form the a-PTC) by footprinting,7,8 lead cleavage,9,10 hydrodynamics, and other biophysical methods, (ii) characterize the enzymatic activity of the a-PTC by the “fragment assay”,11,12 and ultimately, (iii) determine the 3D structure of the aPTC by X-ray diffraction. We are assuming that our first version of the a-PTC will not be enzymatically competent. We will tune it by in vitro selection and other approaches.
1. Hsiao, C. & Williams, L. D. (2009). A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res, 37, 3134-3142.
2. Selmer, M., Dunham, C. M., Murphy, F. V. t., Weixlbaumer, A., Petry, S., Kelley, A. C., Weir, J. R. & Ramakrishnan, V. (2006). Structure of the 70s Ribosome Complexed with mRNA and tRNA. Science, 313, 1935-1942.
3. Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000). The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science, 289, 905-920.
4. Hsiao, C., Mohan, S., Kalahar, B. K. & Williams, L. D. (2009). Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Mol Biol Evol, 26, 2415-2425.
5. Mears, J. A., Sharma, M. R., Gutell, R. R., McCook, A. S., Richardson, P. E., Caulfield, T. R., Agrawal, R. K. & Harvey, S. C. (2006). A Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome. J Mol Biol, 358, 193-212.
6. Bokov, K. & Steinberg, S. V. (2009). A Hierarchical Model for Evolution of 23S Ribosomal RNA. Nature, 457, 977-980.
7. Latham, J. A. & Cech, T. R. (1989). Defining the inside and Outside of a Catalytic RNA Molecule. Science, 245, 276-282.
8. Stern, S., Moazed, D. & Noller, H. F. (1988). Structural Analysis of RNA Using Chemical and Enzymatic Probing Monitored by Primer Extension. Methods Enzymol, 164, 481-489.
9. Behlen, L. S., Sampson, J. R., DiRenzo, A. B. & Uhlenbeck, O. C. (1990). Lead-Catalyzed Cleavage of Yeast tRNAphe Mutants. Biochemistry, 29, 2515-2523.
10. Winter, D., Polacek, N., Halama, I., Streicher, B. & Barta, A. (1997). Lead-Catalysed Specific Cleavage of Ribosomal RNAs. Nucleic Acids Res, 25, 1817-1824.
11. Schmeing, T. M., Seila, A. C., Hansen, J. L., Freeborn, B., Soukup, J. K., Scaringe, S. A., Strobel, S. A., Moore, P. B. & Steitz, T. A. (2002). A Pre-Translocational Intermediate in Protein Synthesis Observed in Crystals of Enzymatically Active 50S Subunits. Nat Struct Biol, 9, 225-230.
12. Anderson, R. M., Kwon, M. & Strobel, S. A. (2007). Toward Ribosomal RNA Catalytic Activity in the Absence of Protein. J Mol Evol, 64, 472-483.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Chiaolong Hsiao
Postdoc
Jessica Bowman
Research Staff
Michelle Williams
Research Staff
Ava Afshar
Undergraduate Student
Jason Murray
Undergraduate Student
Catherine Trippe
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules