2014 Annual Science Report
 University of Illinois at Urbana-Champaign
Reporting | SEP 2013 – DEC 2014
University of Illinois at Urbana-Champaign
Reporting | SEP 2013 – DEC 2014
Project 9: Evolution Through the Lens of Codon Usage
Project Summary
The sequences of protein encoding genes are subject to multiple levels of selection. First, amino acid changes that adversely alter protein function are unlikely to survive. In addition, the genetic code of organisms is degenerate; it includes alternative (synonymous) codons for most of the amino acids. Codon usages in a genome are generally viewed as a balance between drift and selection for rapid and accurate translation of mRNAs into proteins. This balance defines the native codon usage of the organism. Later, it was recognized that many horizontally transferred genes have distinctive codon usages. It was assumed that these reflected the codon usages of the organisms that contributed the genes. We viewed this as an opportunity to identify those sources.
These studies of this have led us to discover that: (i) most of the recently acquired genes come from such closely related organisms that their distinctive codon usages cannot be attributed to a phylogenetically distant source; (ii) the transfers commonly exceed recognized boundaries of microbial species; (iii) after their acquisition, some of the genes do not drift to match the native codon usage of the recipient; (iv) many of the genes that are most up-regulated under starvation conditions have this same codon usage; and (v) a distinctive stress/starvation-associated codon usage is a recurring theme that is observed in diverse Bacteria and Archaea.
These studies entailed the development of a variety of new codon usage analysis tools. We are making these tools available, and are integrating them into the RAST genome annotation and analysis server at Argonne National Laboratory.
Project Progress
9A Evolution through the lens of codon usage: frequent horizontal gene transfer extending beyond species. Over 30 years ago, the codon usages of genes in a genome were interpreted in terms of (i) typical codon usage, (ii) codon usage of abundant proteins (adapted to higher expression levels), and (iii) other maladapted codon usages. In 1991, Médigue et al. recognized that many of these maladapted Escherichia coli genes define a third distinct codon usage type, and noted that many of these genes had been acquired by horizontal gene transfer (i.e., they are alien genes). It has been assumed that the distinctive codon usages reflect those of phylogenetically distant donors, and that with time the alien codon usage of an acquired gene would drift (ameliorate) to match those of the host organism. Using comparative genome analysis, we demonstrated that (i) the codon usage of the most recently acquired genes are indeed distinct, (ii) their codon usage is found only in the genomes of closely related species (i.e., it is not that of a hypothetical distantly related species), (iii) the recently acquired gene codon usages are so similar between E. coli and Salmonella enterica that these species must be drawing upon the same pool of donor genomes, and (iv) that, taken together, these and other closely related species must constitute the reservoir from which the plurality of their recently acquired genes are drawn. This is largely consistent with recent perspectives on the pangenome of species, but it requires that frequent sharing extend substantially beyond current species circumscriptions, which led us to propose a supraspecies pangenome). Neither the pangenome hypothesis, nor our extension of it, provides an explanation for the distinctive codon usage of the recently acquired genes; if they are moving amongst closely related genomes they would be expected to drift to match some average codon usage of those genomes, but they do not.
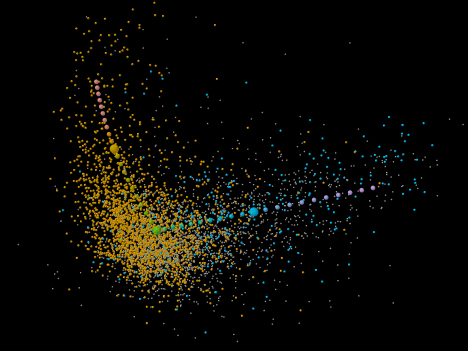
Figure 1. Codon usages of the most recently acquired genes are distinct from those of vertically inherited genes. The illustration shows a factorial correspondence analysis of Salmonella enterica genes separated by differences in codon usage. The first and second axes are shown horizontally and vertically, respectively. The codon usages of vertically inherited genes (orange) form a rabbit head (typical genes) and a left ear (high-expression-adapted genes). These genes can be used to define an expression-level-associated codon usage axis (larger spheres grading from green to red). Recently acquired genes (cyan) have what was originally thought to be alien — but we now consider to be starvation-associated — codon usages, and form a right ear. Starting at the head, this defines a starvation related codon usage axis (larger spheres grading from green to purple). All genes with more complex evolutionary histories are shown in gray.
9B Evolution through the lens of codon usage: vertically inherited genes resist amelioration. The codon usage originally recognized due to its occurrence in recently acquired genes can also be found in genes that have been present in genomes for many 10’s of millions of years. For example, the major “Salmonella Pathogenicity Islands” (SPIs) of S. enterica have been stably present in the genomes since the origins of this species, and some even longer. It had been previously noted that in spite of this, they do not match the G+C content or codon usages of typical or highly expressed genes of the genome. If these genes have been vertically inherited throughout the history S. enterica, they would be expected to drift to match the properties of the rest of the genome, but they have not. Further, we have demonstrated that their codon usages are the same as those of the most recently acquired genes, suggesting a common basis for their distinctive properties.
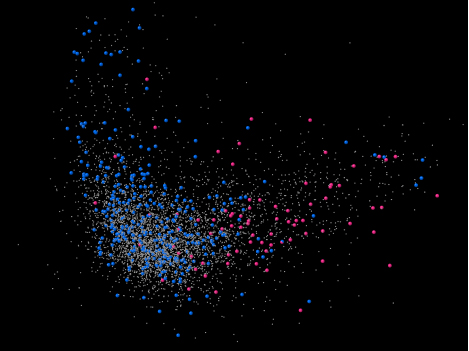
Figure 2. The codon usages of the genes that are most up-regulated under starvation conditions are the same as those of the recently acquired genes. The illustration is the same as Figure 1, but with different genes highlighted. The codon usages of genes whose expression is decreased ≥10-fold by stringent response (starvation signal) in early stationary phase (blue) are mostly along the expression related codon usage axis (head and left ear). The codon usages of the genes whose expression is increased ≥10-fold by stringent response in early stationary phase (pink) are largely distributed along the starvation-associated codon usage axis. All other genes are shown in gray.
9C Evolution through the lens of codon usage: alien gene codon usage is an intrinsic adaptation to stress. It appears that the recently transferred genes and the SPIs are subject to some selective force that acts upon their codon usage. We have shown that is not just a consequence of their G+C content, and it is not just moving the opposite direction from high-expression codon usage. It appears that these genes are subject to positive selection for a third category of codon usage. In seeking clues as to the nature of the selection, the most striking data sets relate to the ppGpp and the stringent response. In textbook molecular biology, the stringent response is responsible for a major shutdown of cell biosynthetic machineries in times of starvation. Two published datasets provide gene expression data under stringent response conditions: one in early stationary phase and one in late stationary phase. Although focus is usually on the things that the stringent response shuts down, the genes most up-regulated (≥10-fold) by this starvation induced response also have the codon use of the SPIs and the recently transferred genes. These data are not fully independent in that the stringent response does induce some of the SPIs. However, stress, and possibly more specifically starvation, are recurring themes when examining the biology of the genes of these codon usages. A starvation codon usage has been previously proposed in the literature, though generally from a more theoretical perspective, and some aspects of our data differ from their predictions.
9D Evolution through the lens of codon usage: starvation codon usage is recurring in nature. We have not completed comprehensive analyses, but many to most of the available genome sequences for Bacteria and Archaea show evidence of a third distinct codon usage (in addition to typical and high-expression). Preliminary analyses of the archaeon Sulfolobus islandicus have led to an additional collaborative project between Whitaker and Olsen in which the codon usage of genes involved in cellular responses to stress induced by viral infection are being investigated. We have not yet examined the genomes of Eucarya.
Tools. The computational tools that we have developed for this work are being integrated into the RAST genome annotation server at Argonne National Laboratory.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Scott Dawson
Project Investigator
Katherine Karberg
Co-Investigator
Gary Olsen
Collaborator
Rachel Whitaker
Collaborator
-
RELATED OBJECTIVES:
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments