2014 Annual Science Report
 University of Illinois at Urbana-Champaign
Reporting | SEP 2013 – DEC 2014
University of Illinois at Urbana-Champaign
Reporting | SEP 2013 – DEC 2014
Project 13: Experimental Determination of the Existence of the Darwinian Transition
Project Summary
Life on our planet can be divided into three domains: Archaea, Bacteria and Eukarya. While some genes may be shared among the domains of life, others especially those involved in information processing namely DNA replication, transcription and translation are often unique to a particular domain. It has, therefore, been proposed that the molecular machineries that carry out these processes (replication, transcription and translation) have crossed a so-called Darwinian threshold where the molecular machineries have become gelled and therefore intolerant of new components. This project is examining the Darwinian threshold hypothesis by testing the interchangeability of the components of the DNA replication machinery across the domains of life. Further experiments will examine the capacity of biomolecules involved in translation and transcription to substitute for their counterparts across the domains of life.
Project Progress
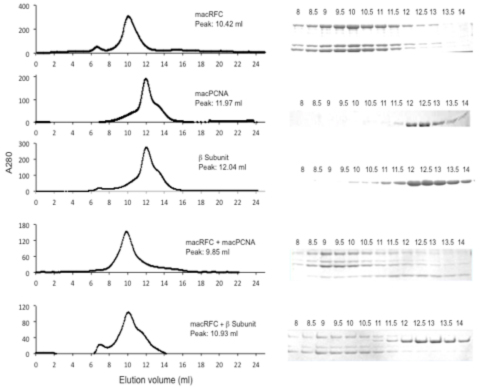
Over the past year we have developed a random transposon insertion library with the goal of identifying conserved hypothetical genes, whose cellular functions are essential. To begin with we developed a genetic marker with strong positive selection for agmatine prototrophy. Using selection for this marker we constructed a transposon mutagenesis system and tested for random insertions by selecting and sequencing insertions. In order to saturate the genome and identify essential genes we estimate that we need to select 20,000 individual random insertions in the 2.59 Mbp genomic background of M.16.4. Great progress has been made toward this goal. We currently have nearly 10,000 clones isolated and estimate that our complete ordered random library will be constructed and sequenced by February 2015.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Isaac Cann
Project Investigator
Hajira Ahmed
Co-Investigator
Carla S.P. Baptista
Co-Investigator
Krunal P. Bodalia
Co-Investigator
Jaigeeth Deveryshetty
Co-Investigator
Matthew Heffernan
Co-Investigator
Satish Nair
Co-Investigator
Miyako Shiraishi
Co-Investigator
Lucy Wan
Co-Investigator
Yoshizumi Ishino
Collaborator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.2
Production of complex life.
Objective 5.3
Biochemical adaptation to extreme environments