2014 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2013 – DEC 2014
Massachusetts Institute of Technology
Reporting | SEP 2013 – DEC 2014
Early Animals: The Genomic Origins of Morphological Complexity
Project Summary
The Peterson lab has continued its focus on micro-RNAs (miRNA) in order to better understand the relationship between genetic and phenotypic diversity. Toward this goal, they have established a new database of miRNA genes (as opposed to sequences) assembled under strict quality control and an entirely novel nomenclature, bridging the different names previously given to the same gene across taxa. This database allows the evolution of miRNA genes to be studied across the animal kingdom. Such study shows that miRNA evolution is not correlated to the duplication of genetic material, but shaped by periods of intense miRNA innovation.
Project Progress
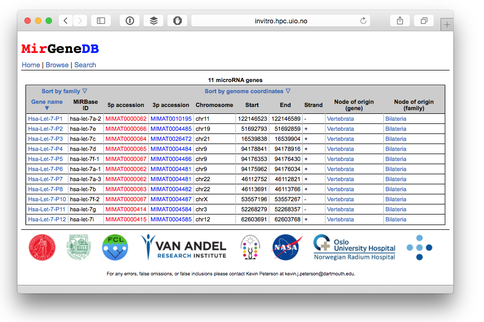
The Peterson lab has continued its focus on microRNAs (miRNA). Because of their importance as gene regulators during animal development, and their relevance in human diseases like cancer, to say nothing of potentially being drivers of morphological complexity, miRNA are among the most intensively studied and discussed molecules of the last 20 years. Although much is known about miRNA procession and structural restraints, their small size (~22 nt), the fact that they are non-coding, and the diversity of miRNA prediction software, determining what is and what is not a miRNA is not straight forward. An understanding of the evolution of miRNAs can only be achieved though if the number of false-positives is virtually zero, and if a common nomenclature—a catalogue of homologous microRNA genes of relevant organisms—is in place. Here we present a uniform system for the annotation and nomenclature of miRNA genes that we apply to the human miRNA entries in miRBase. We show that less than a third of these entries can be supported as derived from canonical miRNA genes, and that the evolutionary history of these miRNA genes was shaped in particular by periods of intense miRNA innovation, periods of evolution not intimately associated with genome duplication events. Further, we show that the mature miRNA gene products have a very different tempo and mode of sequence evolution as compared to the star products, differences related to interactions between mature miRNAs and target mRNAs as well as the star arm and the processing machinery. We establish a new open-access database—MirGeneDB.org (see Fig. 1)—to catalogue this bona fide set of human miRNAs that complements the efforts of miRBase, but differs from it in several fundamental ways including the annotation of mature versus star products, and the imposition of a taxonomic hierarchy upon this curated and consistently-named repertoire.
Publications
-
Field, D. J., Gauthier, J. A., King, B. L., Pisani, D., Lyson, T. R., & Peterson, K. J. (2014). Toward consilience in reptile phylogeny: miRNAs support an archosaur, not lepidosaur, affinity for turtles. Evolution & Development, 16(4), 189–196. doi:10.1111/ede.12081
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Kevin Peterson
Project Investigator
Ben King
Co-Investigator
Davide Pisani
Co-Investigator
Albert Poustka
Co-Investigator
Andreas Schmidt-Rhaesa
Co-Investigator
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.