2013 Annual Science Report
 University of Illinois at Urbana-Champaign
Reporting | SEP 2012 – AUG 2013
University of Illinois at Urbana-Champaign
Reporting | SEP 2012 – AUG 2013
The Nature of the Last Archaeal and Eukaryal Ancestor
Project Summary
The evolutionary history of the eukaryotic cell is intimately linked evolution of atmospheric oxygen and with the endosymbiosis of bacterial symbionts to become the mitochondrial organelles. This project seeks to understand the evolutionary history of the eukaryotic cell using contemporary analogs of ancestral anaerobic eukaryotes (rumen ciliates), which are often associated with endosymbiotic archaea and bacteria in tightly associated communities. We study the evolution of this association using state-of-the-art metagenomic and ecological methods to gain a better understanding of the evolution of these types of associations and thus of eukaryotic evolutionary history.
Project Progress
3.2-1
1. To identify the architecture of the Archaeal genome in which some regions are highly exposed to gene flow and demonstrate communal dynamics where others do not. Variation in recombination rates across chromosomes has been shown to be a primary force shaping the architecture of genome divergence. In archaea, little is known about variation in recombination across the chromosome or how it shapes genome evolution. We identified significant variations in polymorphism occurring across the chromosomes of ten closely-related sympatric strains of the thermoacidophilic archaeon Sulfolobus islandicus. Statistical analyses show that recombination varies across the genome and interacts with selection to define large genomic regions with reduced polymorphism, particularly in the regions surrounding the three origins of replication. Our findings demonstrate how recombination defines the mosaic of variation in this asexually reproducing microorganism and provide insight into the evolutionary origins of genome architecture in this organism from the Archaeal domain.
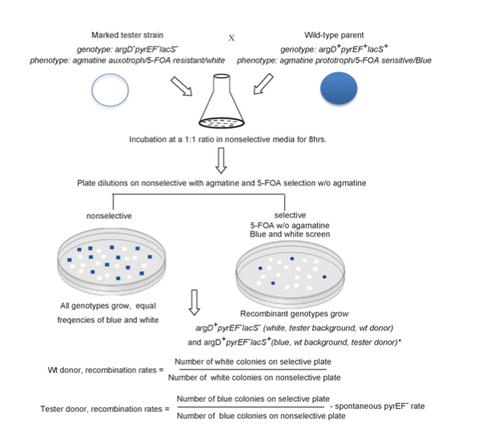
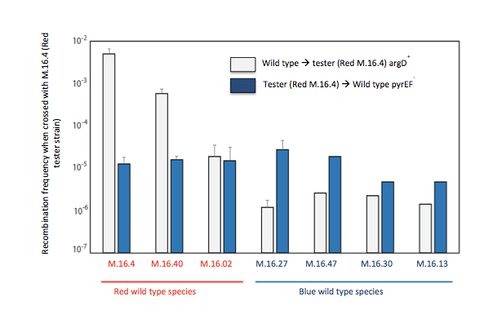
2. Genetic elements play a strong role in the communal dynamics among genomes. They can be easily be added or subtracted from the standing complement of genetic traits, sometimes they are extremely useful, sometimes they contribute little and are just passing through. We have shown the function of genetic elements in speciation in Sulfolobus islandicus. We have developed a high through put system for identifying rates and directionality of gene flow in Sulfolobus islandicus (Figure 1). Based on presence of apparent asymmetry we hypothesize that the mechanism of gene transfer in Sulfolobus is one of conjugation (Figure 2). The intriguing correlation between patterns of matches between the CRISPR adaptive immune loci and elements integrated in microbial genomes, has led us to hypothesize that immune incompatibility could cause isolation between genomes and lead to individualized evolution. We are testing this hypothesis now with genetic knockouts of the CRISPR-cas system in model strains.
3. In order to test the function of annotated genes in the genomes of model archaea and to uncover the function of conserved hypothetical genes in recombination, repair and replication we have developed a robust genetic system to function in the model archaea Sulfolobus islandicus. We are using these markers to specifically knock out genes that we hypothesize are essential to defining chromosome architecture in Archaea such as the RadA homologs present in these genomes as well as replication origins. In addition we will use these markers in a forward genetic screen for novel genes involved in replication, recombination and repair. The first is the agmatine marker described in paper published below. Additionally, 6-methylpurine-resistance determinant, hpt encoded by hypoxanthine phosphoribosyltransferase is identified via both forward and reverse genetic techniques. Strains with lesions in the hpt gene were shown to be > 7.5 fold more resistant to the toxic purine analog 6-methylpurine than as wild type, providing a second counter-selectable marker in S. islandicus.
3.2-2
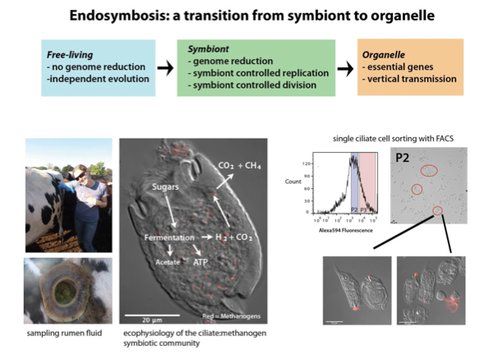
Early eukaryotes and likely evolved low oxygen environments in long-term associations with anaerobic archaea and bacteria. Contemporary anaerobic microbial eukaryotes lacking mitochondria still persist, and a common reservoir for these organisms is the bovine rumen. Anaerobic protists in the rumen ecosystem act as self-contained microbial communities with intimately associated endo- and ectosymbiotic bacteria and archaea. During ciliate fermentation of starch, CO2 and H2 are liberated, promoting syntrophic associations with the presumably mutualistic H2/CO2 consuming archaeal methanogens. In fact, ciliated and flagellated protists comprise over half of the bovine rumen microbial biomass, contributing significantly to degradation of plant material by the concomitant generation the greenhouse gases methane and carbon dioxide by their associations with archaeal symbionts. The interplay of genomes and ecophysiology of these associations are suggestive of eukaryotic evolution and ecology in the anoxic world of early Earth.
Endosymbionts are subject to specific selective pressures, and rumen ciliates likely have a permanent obligate mutualism with endosymbiotic methanogens, resulting in symbiont genome reduction. Similar evolutionary and ecological interactions may have shaped the ecophysiology and evolution of eukaryotic organelles. Thus, we have defined the nature and specificity of the ciliatemethanogen symbiosis through metagenomic comparative and population genomic analyses of a ciliate-methanogen consortium. To characterize the genomes and functions of the rumen ciliate archaeal symbionts, we used Fluorescence-Activated Cell Sorting (FACS) to isolate a community of ciliates with endosymbiotic methanogens from bulk bovine rumen fluid for direct metagenomic sequencing using Illumina MiSeq and HiSeq platforms.
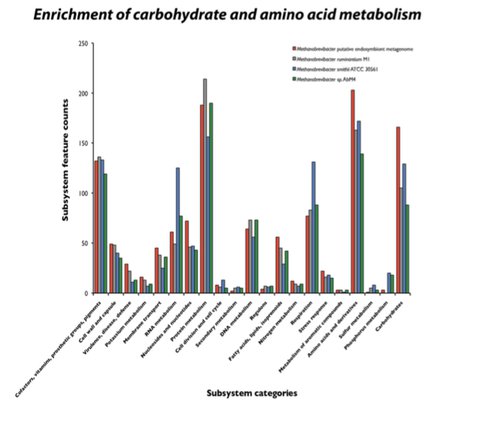
Contigs for endosymbiotic methanogens were assembled using MIRA and Tetra-nucleotide Frequency (TNF) binning, and draft genome assemblies translated and annotated using Interpro. The ciliate archaeal endosymbionts are likely obligate symbionts undergoing purifying selection and possessing reduced genome synteny with increased accumulation of pseudogenes as compared to free-living methanogens. The symbiont genomes are enriched in carbohydrate and amino acid metabolism subsystems (insert Figure 2) yet may have lost specific metabolic pathways, supporting that idea that they may be on an evolutionary path toward be-coming organelles.
We suggest that genomes of early anaerobic eukaryotes were shaped by horizontal gene transfer from similar intimately associated hydrogen-consuming archaea and bacteria. Despite predictions, there has been no direct evidence confirming syntrophic “interspecies hydrogen transfer” (IHT) in this symbiotic association. This work also faciliates an understanding of crucial metabolic processes involved in ciliate-associated methanogenesis to identify pathways that could be targeted to reduce methane emissions.
In the coming year, we also aim to visualize carbon and hydrogen cycling within the ciliatemethanogen symbiosis using the uptake of 13C and 2H at the ciliate sub-cellular level using Transmission Electron Microscopy (TEM) and Nano-scale Secondary Ion Mass Spectrometry (NanoSIMs). The combined use metagenomics, TEM, NanoSIMs are a novel approaches for exploring in situ metabolic transfer and the ecophysiology of a protistan symbiosis.
Publications
-
Krause, D. J., Didelot, X., Cadillo-Quiroz, H., & Whitaker, R. J. (2014). Recombination Shapes Genome Architecture in an Organism from the Archaeal Domain. Genome Biology and Evolution, 6(1), 170–178. doi:10.1093/gbe/evu003
-
Zhang, C., Cooper, T. E., Krause, D. J., & Whitaker, R. J. (2013). Augmenting the Genetic Toolbox for Sulfolobus islandicus with a Stringent Positive Selectable Marker for Agmatine Prototrophy. Applied and Environmental Microbiology, 79(18), 5539–5549. doi:10.1128/aem.01608-13
-
Zhang, C., Krause, D. J., & Whitaker, R. J. (2013). Sulfolobus islandicus: a model system for evolutionary genomics. Biochemical Society Transactions, 41(1), 458–462. doi:10.1042/bst20120338
- Zhang, C., Krause, D.J., Cadiool-Quiroz, H. & Whitaker, R.J. (In Preparation). Heterozygote autoimmunity promotes speciation. Archaea.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Marissa Hirst
Co-Investigator
Kimberly Allen
Collaborator
Eric Estuardo De Guevara
Collaborator
Changyi Zhang
Collaborator
-
RELATED OBJECTIVES:
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.
Objective 5.2
Co-evolution of microbial communities
Objective 6.1
Effects of environmental changes on microbial ecosystems