2013 Annual Science Report
 University of Illinois at Urbana-Champaign
Reporting | SEP 2012 – AUG 2013
University of Illinois at Urbana-Champaign
Reporting | SEP 2012 – AUG 2013
Mining Archaeal Genomes for Signatures of Very Early Life
Project Summary
Carl Woese proposed that life started as semi-autonomous subcellular forms named progenotes. The progenotes lacked cell membranes and readily exchanged information, suggesting that aspects of information processing had already been developed. Woese further hypothesized that certain early life processes crossed a Darwinian threshold, where incorporation of new components of a processes was not tolerated. We aim at determining whether translation, transcription, and replication have crossed the Darwinian threshold. To determine whether DNA replication has crossed the Darwinian Threshold, interchangeability of the DNA replication processivity factor known as the sliding clamp is being examined. It is only in the presence of the sliding clamp that DNA polymerases in extant organisms can gain the speed required to replicate their genomes. In Bacteria, the sliding clamp is the b-subunit of Pol-III and in Archaea and Eukarya the functional homolog is proliferating cell nuclear anti-gen (PCNA). We have, therefore, expressed and purified a sliding clamp from each of the three domains of life (E. coli beta-subunit, M. acetivorans PCNA, and human PCNA). Sliding clamps are loaded in a clamp loader dependent manner; therefore, we have cloned, expressed and purified an archaeal clamp loader from M. acetivorans. Our next step is to determine whether an archaeal clamp loader can interact with each of the sliding clamps from the three domains of life and whether any of the interactions leads to loading of the sliding clamps onto DNA to orchestrate processive DNA synthesis.
Project Progress
*3.1 Interchangeability of sliding clamps from three domains of life *
Sliding clamps are multimeric proteins that promote processivity of DNA polymerases during DNA replication. Sliding clamps encircle DNA and bind to DNA polymerase through a unique peptide present in sliding clamp-interacting proteins. The association of the DNA polymerase with the loaded clamp prevents the frequent dissociations associated with DNA polymerases during DNA synthesis and thus lead to rapid replication of the genome. The sliding clamp is, therefore, central to the capacity to maintain a large genome, a feature common to extant organisms on our planet. To test Carl Woese’s Darwinian threshold hypothesis we have designed an initial experiment that tests interaction of sliding clamps from the three domains of life Bacteria (beta-subunit), Archaea (PCNA) and eukaryotes (PCNA) with the clamp loaders from the different domains.
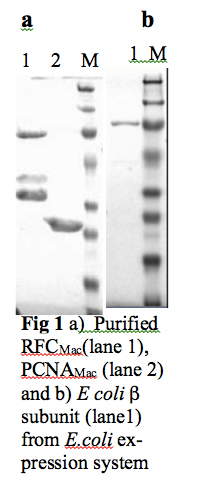
To perform ITC analyses, previously established expression and purification protocols in our lab for archaeal clamp loader RFC (RFCMac) and PCNA (PCNAMac) from Methanosarcina acetivorans were scaled up. Subsequently, RFCMac and PCNAMac were purified to near homogeneity as observed on SDS-PAGE (Fig 1a lane1 and lane 2). E coli β subunit and Human PCNA (PCNAHsa) were cloned and overexpressed in E.coli BL21 (DE3) cells. E coli β subunit was purified to homogeneity by employing Ni NTA and Heparin chromatography columns (Fig. 1 b lane 1). However, PCNAHsa is only partially purified by employing Ni NTA and Heparin chromatography columns. Work is underway to purify PCNAHsa to homogeneity.
RecJ like protein from Methanosarcina acetivorans
RecJ, a bacterial 5’-3’ exonuclease and deoxyribose phosphatase, participates in the DNA repair pathways. RecJ belongs to an exonuclease superfamily with conserved DHH motif. Orthologs of RecJ are found in almost all bacterial species and archaea, but no RecJ homologs are identified in eukaryotes. However, eukaryotic CDC45 belongs to DHH superfamily and interacts with GINS complex, which is also a central nexus in archaeal replication fork. Based on limited sequence similarity to E coli RecJ, two single stranded DNA nucleases were identified in Methanosarcina acetivorans as RecJ-like proteins.
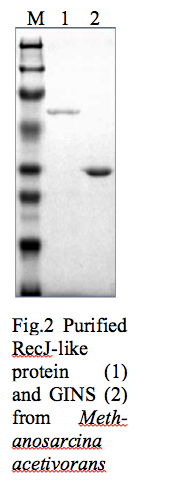
To understand the function and the role of these RecJ-like proteins in Methanosarcina acetivorans cloning of both RecJ-like proteins was carried out. One of the RecJ-like proteins is successfully cloned and expressed in E.coli BL21 (DE3) cells. After identifying optimal expression conditions, RecJ-like protein was purified by using Ni-NTA and heparin columns (Fig. 2 lane1). Biochemical assays will be performed to identify the nuclease activity, substrate and co-factor for this protein. Further, interaction of RecJ-like protein with GINS complex (Fig. 2 lane 2) will be analyzed by gel filtration chromatography. Preliminary data suggests that RecJ-like protein is a nuclease. The second RecJ-like protein identified in Methanosarcina acetivorans is currently cloned.
DNA Polymerase DP1 and DP2
Availability of genomic sequences from archaea reveals that archaeal replication proteins are more closely related to eukaryotic homologs than the bacterial ones. A distinct DNA polymerase (PolD), consisting of a small subunit (DP1) and a large subunit (DP2), is highly conserved in euryarchaeotes and recent genomic data from newly discovered archaeal subdomains show that this new polymerase is only absent in hyperthermophilic crenarchaeotes. DP1 exhibits considerable similarity with the second subunit of eukaryotic DNA polymerase delta while DP2 has conserved motifs found in nucleotide-polymerizing proteins and has some similarity with eukaryotic Pol epsilon. PolD is suggested to replicate the genomes of euryarchaeaote along with a second DNA polymerase known as PolB.
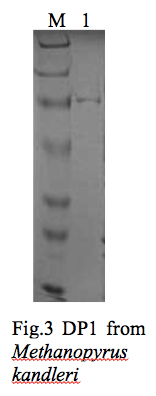
To determine whether this unique DNA polymerase (PolD) can function with DNA replication proteins of other domains of life, the DP1 and DP2 subunits of the hyperthermophilic archaeon Methanopyrus kandleri (Mka) have been cloned. The genes encoding MkaDP1 and MkaDP2 were cloned individually into pET28 and pACYC (mcs2) vectors, respectively. Also, MkaDP1 and MkaDP2 were cloned into pACYC duet (mcs1 and mcs2 respectively) to examine the formation of DP1 and DP2 complex. Currently only DP1 has been expressed and purified using Talon and Heparin chromatography columns, although the yields have been very low (Fig. 3). We are making modifications to our gene expression system to increase yield of the two proteins during expression in E. coli.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Isaac Cann
Project Investigator
Zaida Luthey-Schulten
Co-Investigator
Jaigeeth Deveryshetty
Collaborator
Yoshizumi Ishino
Collaborator
Jennifer McGinnis
Collaborator
Kelly Mehan
Collaborator
Noor Soufan
Collaborator
Lucy Wan
Collaborator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.2
Production of complex life.