2012 Annual Science Report
 Pennsylvania State University
Reporting | SEP 2011 – AUG 2012
Pennsylvania State University
Reporting | SEP 2011 – AUG 2012
Developing New Biosignatures
Project Summary
The Developing New Biosignatures project is aimed at creating innovative approaches for the analyses of cells and other organic material, finding ways in which metal abundances and isotope systems reflect life, and developing creative approaches for using environmental DNA to study present and past life.
Project Progress
Fossil preservation in Miocene and Permian sulfates:
By use of optical microscopy, confocal laser scanning microscopy, and Raman spectroscopy, we have documented the presence of diverse fossilized microorganisms in Miocene deposits of Italy and Permian gypsums of Texas and New Mexico. As recently reported (2012, Astrobiology 12: 619-633), these findings, coupled with our discovery of gypsum-permineralized microscopic fossils in Recent gypsum deposits of Mexico, Peru, and Australia, have obvious relevance to the search for evidence of past life on Mars where deposits of sulfate, including gypsum, are abundant and widespread.
Chert-permineralized Duck Creek (~1,800 Ma) sulfuretum from Western Australia:
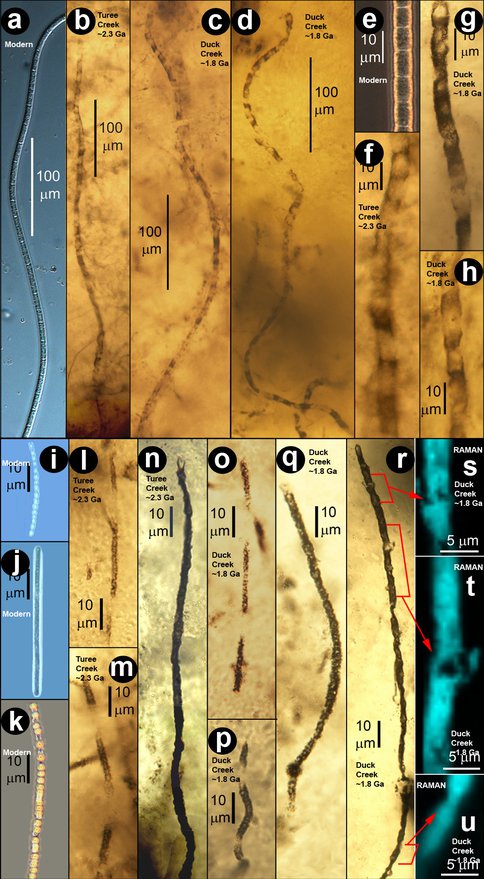
Our PSARC summary of 2011 reported discovery of a fossilized non-photosynthetic ecosystem previously unknown from the geological record: the sub-sediment sulfur-cycling microbiota of the 2,300-Ma-old Turee Creek chert of Australia (VanKranedonk et al., Submitted). We have now discovered a second example, ~500 Ma younger, permineralized in cherts of the ~1,800-Ma-old Duck Creek Fm., also of Western Australia. Significantly, the two biotas are essentially identical — in ecologic setting, cobweb-like non-stromatolitic fabric, and microbial composition. These two microbial assemblages, and their striking similarity in all respects to a modern sub-seafloor sulfuretum off the coast of Chile (Figure 1), establish for the first time the prime null hypothesis required by Darwin’s theory of evolution: if there is no change in the physical/biological environment, there can be no evolution.
Figure 1. Comparison of modern (a, e, i-k), ~2.3 Ga Turee Creek (b, f, l-n), and ~1.8 Ga Duck Creek (c, d, g, h, o-u) broad (a-h) and “bead-cell” (i-u) filamentous sulfuretum microbes; (s-u) show Raman images of the filament in®, acquired at the ~1605 cm-1 band of kerogen, that establish its carbonaceous composition.
Scytonemin:
Many species of cyanobacteria tolerate exposure to harmful levels of ultraviolet (UV) radiation by producing photoprotective pigments. Scytonemin, found in extracellular polysaccharide sheaths, is produced in abundance when terrestrial or benthic cyanobacteria are exposed to direct sunlight, such as in desertsoil crusts and intertidal mats. It is exclusive to cyanobacteria and therefore can serve as a diagnostic biomarker, particularly for UV-exposure growth conditions. We have shown that it is preserved in abundance in mid-Holocene sedimentary intervals in the BlackSea (Fulton et al., 2012), a novel deep sea occurrence that demonstrates that scytonemin is resistant to degradation during erosion and transport. C and N isotopic compositions support the interpretation that scytonemin was derived from cyanobacteria in cryptobiotic desertsoil, suggestive of expanding aridity during the Subboreal Phase in the BlackSea region. Scytonemin has potential for preservation in black shales, where it may serve as an important biomarker for tracing the evolution and expansion of cyanobacterial populations, especially in association with elevated UV stress.
Coenzyme F430:
Methyl-coenzyme M reductase, an enzyme traditionally associated with methanogenesis, has recently been linked to the anaerobic oxidation of methane suggesting methane oxidation follows a pathway similar to reverse methanogenesis. Coenzyme F430, a tetrapyrrole-nickel complex within the active site of methyl-coenzyme M, is used in methanogenesis and is hypothesized to play a key role in archaeal methanotrophy (Scheller et al., 2010). We recently developed a method to extract and isolate F430 from natural sediments so it can be purified for carbon and nitrogen stable isotope analysis. We have produced concentration and isotopic data for coenzyme F430 from active seep sediment cores from the Eel River Basin (California), a site where the anoxic oxidation of methane occurs. A spike in the concentration of F430 is observed at the 3-6 cm depth horizon corresponding with peak abundance in ANME-2/Desulfosarcina/Desulfococcus aggregate counts. Carbon isotope values of F430 are significantly more enriched in 13C (-23‰ to -26‰) than published diphytanyl glycerol diether archaeal lipids and cell clusters measured with nano-SIMS. This unexpected observation suggests that F430 in this methanotrophic sediment may not be uniquely associated with methanotrophs and that methanotrophy may not take place via the reversal of methanogenesis. However, the enriched signal reported here could also be due to another carbon substrate for the ANME-2. The relatively 13C-enriched F430 will guide further exploration of the pathways of carbon cycling in these marine sediments.
Tintinnids:
Tintinnid are small microfossils found in the 715-635 Ma Tayshir member of the Tsagaan Oloom Formation, Mongoli. It is belived that these tintinnids represent the organic matter production in the surface waters. Samples of 100 to 150 tintinnids were sent to us at penn state from the Bosak lab at MIT for carbon and nitrogen isotope analysis. Using our Nano elemental analyzer isotope ration mass spectrometer were are able to make carbon and nitrogen isotope measurements at the nano molar scale required. We find a relatively homogeneous carbon isotope pattern through the Tayshir member that is isotopically distinct from the carbonate. This works will be presented as the upcoming AGU Fall 2012 meeting.
Polyphosphate as a biosignature:
Results have confirmed previous preliminary results that stored polyphosphate supports growth in the absence of methanogenesis. Conditional mutants have been constructed in which the polyphosphate kinase 2 (PPK2) gene is deleted.
Secondary Ion Mass Spectrometry (SIMS) development – Carbon isotopic analysis
A paper was published where we applied our FISH and FISH-SIMS analyses using 13C and 15N labeled substrates to microbes living in methane seep environments, we find that the most active cells during manganese dependent anaerobic methane oxidation are primarily mixed and mixed-cluster aggregates of archaea and bacteria. Overall, our control experiment using sulfate showed two active bacterial clusters, two active shell aggregates, one active mixed aggregate, and an active archaeal sarcina, the last of which appeared to take up methane in the absence of a closely-associated bacterial partner. A single example of a shell aggregate appeared to be active in the manganese incubation, along with three mixed aggregates and an archaeal sarcina. These results suggest that the microorganisms (e.g., ANME-2) found active in the manganese-dependent incubations are likely capable of sulfate-dependent anaerobic oxidation of methane.
We also applied SIMS to possible fossil cells from the Archean. Interpretation of organic microstructures in the ~3 Ga Farrel Quartzite (FQ) of Australia has been challenging because of the surprisingly large size and unusual spindle-like morphology of some of the constituent forms. We have submitted a paper reporting 33 in-situ carbon isotopic analyses of 16 individual FQ specimens. The results provide new insights into the biogenicity and evolutionary significance of these microstructures. Our data demonstrate that both the spheroids and spindles of the FQ have mean δ13C values of 37.1±0.2‰, an isotopic composition that is markedly distinct from that determined for the background organic matter in the same thin section (mean δ13C = 32.9±0.4‰). This isotopic distinction shows that the preserved microstructures are not pseudofossils (potentially formed from physical reprocessing of the bulk sedimentary organic material), and when considered along with published morphological and chemical studies, argues that the FQ spindles and spheroids are bona fide microfossils. Our results also provide metabolic constraints that indicate most of these preserved microorganisms were autotrophic (with a few sphereoids possibly being methanotrophic). Finally, the isotopic data suggest that the fossils were transported into the sedimentary environment and therefore support an interpretation that the spindles represent the remains of planktonic microorganisms. The existence of similar spindles in the ~3.4 Ga Strelley Pool Formation of Australia and the ~3.4 Ga Onverwacht Group of South Africa suggests that the spindle-containing microbiota may be one of the oldest, morphologically preserved examples of life on Earth. If this is the case, then the FQ structures may well be remains of a cosmopolitan biological experiment that appears to have lasted for several hundred million years, starting in the Paleoarchean. A paper on this work has been submitted for publication.
SIMS development – Metal Analyses:
As part of a NASA Astrobiology Postdoctoral Fellowship to Jennifer Glass through PSARC, the Caltech team has been working to develop methods to image the intercellular distribution of metals in symbiotic aggregates of sulfate-reducing bacteria (SRB) and anaerobic methane-oxidizing archaea (ANME) using nanoSIMS. The overarching goal of this work is to determine whether differences in cellular metal concentration exist between SRB and ANME. Hydrate Ridge sediment was incubated with addition of isotopically-labeled metals (54Fe, 57Fe, 92Mo, 95Mo and 60Ni) and samples were preserved at regular intervals for nanoSIMS analysis. Aggregates were separated from sediment by centrifugation in a Percoll density gradient, embedded in LR-white resin, and cut into 0.5-1 mm thin sections. These thin sections were then placed onto a silica wafer and analyzed by nanoSIMS. The first run was performed using the Cs+ beam, which sputters negative ions. We measured the cellular distribution of Fe, the metal likely to be the most abundant in the aggregates, and its negative molecular ions FeC, FeO and FeS, in aggregates with and without a clay crust. We found that of the molecular ions mapped, FeO gave the strongest signal. A clear ring of Fe was visible around crusted aggregates, whereas uncrusted aggregates showed minimal Fe. The trade-offs of using the Cs+ beam as opposed to the O2- beam are higher spatial resolution and much higher signal on CN and S, but lower sensitivity for metals since only negatively molecular ions are sputtered. Therefore, the second run was performed using the O2- beam, which sputters positive ions, in order to optimize detection of positively charged metals. In this run we measured the cellular distribution of Fe as well as Ni, likely the second most abundant metal in the cells, in aggregates from an 8-month time point of incubations with and without isotopically labeled 54Fe. Carbon counts were very low (<20 12C counts per second) and therefore sodium (23Na) was used as a cellular indicator instead. 56Fe and 58Ni tended to co-localize in hot spots. Due to the low spatial resolution in O2- mode, it is difficult to determine whether these “hot spots” were actually cells, or if they were instead clay. However, it seems more likely that they represent cellular metals as the “hot spots” also co-occurred with areas where both C and Na were present. Determining whether the Fe and Ni were coming from ANME or SRB was also difficult since only DAPI images were available for comparison. No differences in 56Fe/54Fe were observed between the 54Fe-labeled and unlabeled aggregates; both were not significantly different than the natural abundance ratio of 15.7. It is probable that the high concentrations of Fe available to the cells from the sediment swamped the signal such that minimal 54Fe isotope was incorporated. These results represent a further extension of the analytical capabilities of the nanoSIMS for microbial cell analysis and the first efforts toward localization of intercellular metals in methane-oxidizing ANME/SRB aggregates. Future work will attempt to optimize the spatial resolution and improve upon the detection limits of metals, potentially aided by the analysis of thin sectioned methane-oxidizing aggregates after fluorescence in situ hydridization. Demonstration of the incorporation of isotopically-labeled metals into cellular biomass for these slow growing organisms continues to be a challenge, but may prove successful as a proof of concept using pure cultures of archaea and bacteria.
Weathering as a possible biosignature:
We are studying weathering of rocks of basaltic composition. We are studying interactions between geochemical and biological processes that contribute to rock weathering, colloid formation, and element mobility in basaltic systems. We are comparing temperature versus cold/dry climates. We have discovered that colloid formation is important at these sites and we have been investigating that too. One site is Svalbard, a Mars analog site north of the Arctic Circle, and the other site is a PA diabase near Gettysburg, PA.
DNA as a biosignature:
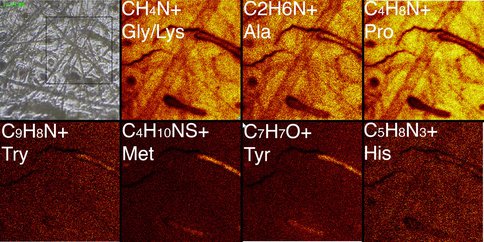
We are attempting to learn the limits of DNA preservation in different environments, focusing on permafrost (up to 700,000 years old), but also including a recently discovered wet, temperate mountain lake site that spans the last interglacial through the peak of the last glacial maximum (ca 150,000 years ago – ca 25,000 years ago). We have collected large mammal bones (horse, mammoth bison) from each site and processed these for DNA amplification, using an in-solution capture protocol with second and third generation sequencing platforms. We have been surprisingly successful in extracting DNA from very old permafrost-preserved specimens. To date, we have complete mitochondrial genomes from several hundred horses ranging in age form 12,000 years to 700,000 years old. One of three 700,000 year old horse bones has also been used for shotgun sequencing of its full genome to 1.5X, using Illumina and Helicos sequencing platforms. This marks the oldest recovered DNA to date, and is the subject of a manuscript that has just been submitted to Nature. Complete mitochondrial genomes and targeted nuclear loci have also been recovered from horses estimated to 80,000, 160,000 and 400,000 years old, based on age estimates from associated volcanic tephra. We have had less success with mammoth and bison bones recovered from the high mountain site (Snowmass, Colorado), but remain optimistic as we wait for the results of our capture experiment. The differences in preservation are most likely due to moisture, in particular the repeated freeze/thaw cycles to which bones in the high altitude rockies are subjected are likely to fragment DNA molecules to the point where they are not normally recoverable. We are performing additional tests, using both DNA and protein detection protocols, with the aim to generate a predictable model for biomolecular survival in different preservation environments. We have recently used Time of Flight Secondary Ion Mass spectrometry to investigate the preservation of peptides in a 700,000 year old horse metapodial recovered from the Klondike Gold Fields, Yukon Territory Canada (Figure 2).
Figure 2. Time of Flight Secondary Ion Mass spectrometry analysis of a 700,000 year old horse metapodial recovered from the Klondike Gold Fields, Yukon Territory Canada, showing abundant secondary ions characteristic of amino-acid peptides. Glycine, proline and alanine are enriched, while other amino acids such as methionine and tyrosine are detected at lower levels, as is typical of collagen. The presence of collagen, together with 72 additional proteins including blood-derived peptides, was confirmed by direct sequencing using high-resolution mass spectrometry proteomics, suggesting excellent biomolecular preservation of the bone.
We also extended work on whole genome amplification of DNA from low biomass deeply-buried sediment samples. We published a method paper describing a degenerate polymerase chain reaction (PCR)-based method of whole-genome amplification. The method is designed to work fluidly with 454 sequencing technology, was developed and tested for use on deep marine subsurface DNA samples. While optimized for use with Roche 454 technology, the general framework presented may be applicable to other next generation sequencing systems as well (e.g., Illumina, Ion Torrent). The method, which we have called random amplification metagenomic PCR (RAMP), involves the use of specific primers from Roche 454 amplicon sequencing, modified by the addition of a degenerate region at the 3′ end. It utilizes a PCR reaction, which resulted in no amplification from blanks, even after 50 cycles of PCR. After efforts to optimize experimental conditions, the method was tested with DNA extracted from cultured E. coli cells, and genome coverage was estimated after sequencing on three different occasions. Coverage did not vary greatly with the different experimental conditions tested, and was around 62% with a sequencing effort equivalent to a theoretical genome coverage of 14.10×. The GC content of the sequenced amplification product was within 2% of the predicted values for this strain of E. coli. The method was also applied to DNA extracted from marine subsurface samples from ODP Leg 201 site 1229 (Peru Margin), and results of a taxonomic analysis revealed microbial communities dominated by Proteobacteria, Chloroflexi, Firmicutes, Euryarchaeota, and Crenarchaeota, among others. These results were similar to those obtained previously for those samples; however, variations in the proportions of taxa identified illustrates well the generally accepted view that community analysis is sensitive to both the amplification technique used and the method of assigning sequences to taxonomic groups. Overall, we find that RAMP represents a valid methodology for amplifying metagenomes from low-biomass samples.
Prebiotic organics:
Graduate student Karen Smith continued work on the organics in meteorites in collaboration with NASA Goddard (Jason Dworking and Michael Callahan). She made great progress with the characterization of nicotinic acid (and its isomers) in a suite of carbonaceous chondrites. She also identified extractable HCN in several meteorites and is working to understand its origin.
Publications
-
Fulton, J. M., Arthur, M. A., & Freeman, K. H. (2012). Subboreal aridity and scytonemin in the Holocene Black Sea. Organic Geochemistry, 49, 47–55. doi:10.1016/j.orggeochem.2012.05.008
-
Fulton, T. L., Wagner, S. M., Fisher, C., & Shapiro, B. (2012). Nuclear DNA from the extinct Passenger Pigeon (Ectopistes migratorius) confirms a single origin of New World pigeons. Annals of Anatomy – Anatomischer Anzeiger, 194(1), 52–57. doi:10.1016/j.aanat.2011.02.017
-
Ginolhac, A., Vilstrup, J., Stenderup, J., Rasmussen, M., Stiller, M., Shapiro, B., … Orlando, L. (2012). Improving the performance of True Single Molecule Sequencing for ancient DNA. BMC Genomics, 13(1), 177. doi:10.1186/1471-2164-13-177
-
Higham, T., Compton, T., Stringer, C., Jacobi, R., Shapiro, B., Trinkaus, E., … Fagan, M. (2011). The earliest evidence for anatomically modern humans in northwestern Europe. Nature, 479(7374), 521–524. doi:10.1038/nature10484
-
House, C. H., Beal, E. J., & Orphan, V. J. (2011). The Apparent Involvement of ANMEs in Mineral Dependent Methane Oxidation, as an Analog for Possible Martian Methanotrophy. Life, 1(1), 19–33. doi:10.3390/life1010019
-
House, C. H., Oehler, D. Z., Sugitani, K., & Mimura, K. (2013). Carbon isotopic analyses of ca. 3.0 Ga microstructures imply planktonic autotrophs inhabited Earth’s early oceans. Geology, 41(6), 651–654. doi:10.1130/g34055.1
-
Kistler, L., & Shapiro, B. (2011). Ancient DNA confirms a local origin of domesticated chenopod in eastern North America. Journal of Archaeological Science, 38(12), 3549–3554. doi:10.1016/j.jas.2011.08.023
-
Letts, B., & Shapiro, B. (2011). Case Study: Ancient DNA Recovered from Pleistocene-Age Remains of a Florida Armadillo. Ancient DNA, None, 87–92. doi:10.1007/978-1-61779-516-9_12
-
Lorenzen, E. D., Nogués-Bravo, D., Orlando, L., Weinstock, J., Binladen, J., Marske, K. A., … Willerslev, E. (2011). Species-specific responses of Late Quaternary megafauna to climate and humans. Nature, 479(7373), 359–364. doi:10.1038/nature10574
-
Martino, A. J., Rhodes, M. E., Biddle, J. F., Brandt, L. D., Tomsho, L. P., & House, C. H. (2012). Novel Degenerate PCR Method for Whole-Genome Amplification Applied to Peru Margin (ODP Leg 201) Subsurface Samples. Frontiers in Microbiology, 3. doi:10.3389/fmicb.2012.00017
-
Ming, R., VanBuren, R., Liu, Y., Yang, M., Han, Y., Li, L-T., … Shen-Miller, J. (2013). Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biology, 14(5), R41. doi:10.1186/gb-2013-14-5-r41
-
Minyard, M. L., Bruns, M. A., Liermann, L. J., Buss, H. L., & Brantley, S. L. (2012). Bacterial Associations with Weathering Minerals at the Regolith-Bedrock Interface, Luquillo Experimental Forest, Puerto Rico. Geomicrobiology Journal, 29(9), 792–803. doi:10.1080/01490451.2011.619640
-
Molak, M., Lorenzen, E. D., Shapiro, B., & Ho, S. Y. W. (2012). Phylogenetic Estimation of Timescales Using Ancient DNA: The Effects of Temporal Sampling Scheme and Uncertainty in Sample Ages. Molecular Biology and Evolution, 30(2), 253–262. doi:10.1093/molbev/mss232
-
Schopf, J. W., & Kudryavtsev, A. B. (2012). Biogenicity of Earth’s earliest fossils: A resolution of the controversy. Gondwana Research, 22(3-4), 761–771. doi:10.1016/j.gr.2012.07.003
-
Schopf, J. W., Farmer, J. D., Foster, I. S., Kudryavtsev, A. B., Gallardo, V. A., & Espinoza, C. (2012). Gypsum-Permineralized Microfossils and Their Relevance to the Search for Life on Mars. Astrobiology, 12(7), 619–633. doi:10.1089/ast.2012.0827
-
Shi, C. S., Schopf, J. W., & Kudryavtsev, A. B. (2013). Characterization of the stem anatomy of the Eocene fern Dennstaedtiopsis aerenchymata (Dennstaedtiaceae) by use of confocal laser scanning microscopy. American Journal of Botany, 100(8), 1626–1640. doi:10.3732/ajb.1300027
-
Williford, K. H., Ushikubo, T., Schopf, J. W., Lepot, K., Kitajima, K., & Valley, J. W. (2013). Preservation and detection of microstructural and taxonomic correlations in the carbon isotopic compositions of individual Precambrian microfossils. Geochimica et Cosmochimica Acta, 104, 165–182. doi:10.1016/j.gca.2012.11.005
- Letts, B., Fulton, T.L., Stiller, M., Andrews, T., MacKay, G., Popkp, R. & Shapiro, B. (2012). Ancient DNA Reveals Genetic Continuity in Mountain Woodland Caribou of the Mackenzie and Selwyn Mountains, Northwest Territories, Canada. Arctic Institute of North America, 65(5).
- Nasdala, L., Beyssac, O., Schopf, J.W. & Bleisteiner, B. (2012). Application of Raman-based images in the Earth sciences. In: Zoubir, A. (Eds.). Optical Sciences. Vol. 168. Raman Imaging.
- Orlando, L., Ginolhac, A., Froese, D., Vilstrup, J., Raghavan, M., Stiller, M., Cappelini, E., Rasmussen, M., Zazula, G., Weinstock, J., Hofreiter, M., Gilbert, M.T.P., Zhang, G., Olsen, J.V., Nielsen, R., Shapiro, B., Jun, W. & Willerslev, E. (2012, Submitted). Horse evolutionary genomics as revealed by a 700,000 year old genome.
- Schopf, J.W. & Kudryavtsev, A.B. (2012, Submitted). Biogenicity of Earth’s earliest fossils. In: Dilek, L. & Furnes, H. (Eds.). Archean Earth and Early Life. Springer.
- Schopf, J.W. (2012). The fossil record of cyanobacteria. In: Whitton, B.A. (Eds.). Ecology of Cyanobacteria II. Their Diversity in Space and Time. Berlin Heidelberg New York: Springer.
- Schopf, J.W. (2012, In Prss). Forward [Book Chapter]. The Flyer: Boris Sokolov: Natural History and 21st Century Russia Sokolov. Indiana University Press.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Tanja Bosak
Collaborator
Jorge Bueno
Collaborator
Nicholas Butterfield
Collaborator
Charles Cockell
Collaborator
George Cody
Collaborator
Phoebe Cohen
Collaborator
Andrew Czaja
Collaborator
Jason Dworkin
Collaborator
Jack Farmer
Collaborator
Duane Froese
Collaborator
Libby Hausrath
Collaborator
Francis Macdonald
Collaborator
Ken Takai
Collaborator
Martin Van Kranendonk
Collaborator
Malcolm Walter
Collaborator
Maria Zambrano
Collaborator
Venkata Vepachedu
Postdoc
John Cantolina
Research Staff
Anatoliy Kudryavtsev
Research Staff
Laura Liermann
Research Staff
Heather Nelson
Research Staff
Dennis Walizer
Research Staff
Yumiko Watanabe
Research Staff
Zhidan Zhang
Research Staff
Michael Avery
Graduate Student
Laurence Bird
Graduate Student
Leah Brandt
Graduate Student
Anne Dekas
Graduate Student
Ian Foster
Graduate Student
Khadouja Harouaka
Graduate Student
Mandi Martino
Graduate Student
Karen Smith
Graduate Student
Tiffany Yesavage
Graduate Student
Danielle Gruen
Undergraduate Student
Muammar Mansor
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 7.1
Biosignatures to be sought in Solar System materials