2012 Annual Science Report
 Pennsylvania State University
Reporting | SEP 2011 – AUG 2012
Pennsylvania State University
Reporting | SEP 2011 – AUG 2012
Biosignatures in Relevant Microbial Ecosystems
Project Summary
PSARC is investigating microbial life in some of Earth’s most mission-relevant modern ecosystems. These environments include the extremely salty Dead Sea, the impact-fractured crust of the Chesapeake Bay impact structure, methane seeps on the ocean floor, deep ice in the Greenland ice sheet, and oxygen-free waters including deep subsurface groundwater. We target environments that, when studied, provide fundamental information that can serve as the basis for future solar system exploration. Combining our expertise in molecular biology, geochemistry, microbiology, and metagenomics, and in collaboration with some of the planet’s most extreme explorers, we are deciphering the microbiology, fossilization processes, and recoverable biosignatures from these mission-relevant environments.
Project Progress
Fossil preservation in recent sulfates
Schopf (UCLA), Kudryavtsev (UCLA), Foster (UCLA) (in collaboration with J. Farmer (ASU), M. Walter (UNSW), V. A. Gallardo (Universidad de Concepcion, Chile), Carola Espinoza (Universidad de Concepcion, Chile)
By use of optical microscopy, confocal laser scanning microscopy, and Raman spectroscopy, we documented the presence of diverse fossilized microorganisms in Recent gypsum deposits of Mexico, Peru, and Australia. As recently reported (2012, Astrobiology 12: 619-633), these findings, coupled with our discovery of gypsum-permineralized microscopic fossils in Miocene deposits of Italy and Permian gypsums of Texas and New Mexico, are relevant to the search for evidence of past life on Mars where deposits of sulfate, including gypsum, are abundant and widespread.
In addition to the UCLA contributors noted above (J. William Schopf, Anatoliy B. Kudryavtsev, and Ian B. Foster), contributors to this research include four NAI-supported US-based scientists: Jack D. Farmer (Arizona State University) and Kenneth H. Williford, Reinhard Kozdon and John W. Valley (University of Wisconsin, Madison) as well as two NAI-supported Australian scientists: Malcolm R. Walter and Martin Van Kranendonk (University of New South Wales, Australia). Moreover, Schopf’s international ties to the Australians noted above and the Chilean scientists involved in this research (Victor A. Gallardo and Carola Espinoza, Universidad de Concepción), as well as his membership on the advisory bodies of the Brazilian Astrobiology Center (Universidade de Sao Paulo) and the Geobio-Center of the Ludwig-Maximilians-Universität (Munich, Germany), would not have occurred had he lacked NAI affiliation.
Our studies of extant microorganisms (biology), fossil microscopic organisms (paleontology), the mineralogy and taphonomy of their preservation (geology), and their chemical composition (geochemistry) are highly interdisciplinary, involving both the life sciences (biology, microbiology, evolutionary biology) and the physical sciences (geology and geochemistry).
The detection by J.W. Schopf et al. of diverse microscopic fossils preserved in gypsum (2012, Astrobiology 12: 619-633) is of significance for the potential discovery of fossils in Mars sulfates and should influence the selection of sites for Mars surface sample collection. This discovery, coupled with that of Schopf and colleagues, including PSARC investigator Macalady, of a previously unreported Precambrian sulfur-dependent ecosystem — sulfureta essentially identical to that living today and now thought to be preserved in 1,800- and 2,300-Ma-old geological units — hold promise for the discovery of ancient and perhaps even extant life on Mars. Macalady’s involvement in the work (in preparation for publication) linking modern and ancient microbial sulfureta is a direct result of NAI activities.
Peer reviewed publications
SCHOPF, J.W., FARMER, J.D., FOSTER, I.S., KUDRYAVTSEV, A.B., GALLARDO, V.A., and ESPINOZA, C. 2012. Gypsum-permineralized microfossils and their relevance to the search for life on Mars. Astrobiology 12: 619-633.
Published Book Chapters
NASDALA, L., BEYSSAC, O., SCHOPF, J.W., and BLEISTEINER, B. 2012. Application of Raman-based images in the Earth sciences. In: A. Zoubir (Ed.), Raman Imaging, Springer Series in Optical Sciences vol. 168 (Amsterdam: Springer), pp. 145-187.
Molecular signatures of microbial life in the Dead Sea House, Freeman, Macalady & graduate students M. Rhodes and K. Dawson (in collaboration with A. Oren & J. R. Spear)
We continued our work on hypersaline environments by comparing rRNA amplicon libraries from a haloarchaeal bloom in the hypersaline Dead Sea in 1992 to the 2007 residual population and simulated blooms in experimental mesocosms. Our high-throughput work on the Dead Sea wrapped up with a journal article showing how the Dead Sea microbial populations shifted during bloom events in 1992 and surprisingly that a signature from the microbial bloom was retained 15 years later. Moshe Rhodes, the graduate student working primarily on this project has now successfully finished his Ph.D.
Graduate student Katherine Dawson’s paper describing the response of archaeal halophile membrane chemistry to salinity levels has been published. She presented strong evidence that the degree of membrane lipid unsaturation is an important adaptation to specific salinity niches in archaeal halophiles. The new findings provide a solid footing from which to unravel the contributions of archaeal membrane physiology and shifts in community composition to the organic biomarker record of salinity fluctuations in the Dead Sea.
The project required geologic understanding of the Dead Sea basin, as well as modern state-of-the-art molecular biological tools. NAI provided a platform for establishing collaborations with Aharon Oren and for an earlier paper with John Spears.
Peer reviewed publications:
Rhodes, M. E., Oren, A., and House, C. H. 2012. Dynamics and persistence of Dead Sea microbial populations as shown by high-throughput sequencing of rRNA. Applied and Environmental Microbiology 78:2489-2492.
Dawson, K. S. , Freeman, K. H., and Macalady, J. L. 2012. Molecular characterization of core lipids from halophilic archaea grown under different salinity conditions. Organic Geochemistry 48:1-8.
Abstracts presented:
Dawson, K. S.*, Schaperdoth, I., Freeman, K. H., and Macalady, J.L. 2012. Anaerobic biodegradation of the isoprenoid biomarker analogues pristane and phytane. Gordon Research Conference: Organic Geochemistry, July 2012.
Impact cratering and subsurface microbial life Macalady, Shapiro & graduate student Korzow-Richter (in collaboration with C. Cockell, D. Vanko & M. Voytek)
Additional personnel: Dr. Mathias Stiller (helped with library preparation of core DNA extracts), Dr. Lisa Steinberg (assisted with DNA methods)
Astrobiology dual title Ph.D. student Kristine Korzow-Richter, co-advised by Beth Shapiro (primary) and Jenn Macalady, continued her work on authenticated core samples from the ~2 km deep Chesapeake Bay impact structure. We compared six DNA extraction protocols, including one developed specifically for this project, to determine the most appropriate method for extracting DNA from the Chesapeake Bay core. Among the most challenging aspects of this project is the extremely low biomass expected within each of the sampled sediment layers. A highly efficient method of DNA recovery is therefore required. Our initial comparisons revealed little if any difference between the six methods, but showed that when biomass was very low, recovery was sporadic at beast.
We therefore performed DNA extraction on sediment sampled from six different depths of the core spanning 159-897 meters below the sediment surface, using each of the six methods. DNA extraction from deep sea sediments was also performed as a positive control. In a few cases. both from the core and from the deep sea sediments, we succeeded in amplifying DNA sequences from the resulting extract using a series of conserved primer pairs. All DNA extracts for which amplification success was observed, plus several positive and negative controls have been prepared for sequencing on the Illumina HiSeq platform, and submitted to UC Berkeley, where they are currently in the queue for sequencing. We are currently awaiting the result. In addition to this work, we are currently generating sequence data from previously reported cultured bacteria from a different layer in the core . These sequence data, generated using Sanger sequencing technology, will be used to test whether the cultured bacteria are authentically ancient (see below).
In addition to the laboratory work described above, we have been developing a computational test to determine the authenticity of any DNA sequence data generated from the ancient cores. As with any ancient DNA project, the possibility exists that the sample was contaminated at some point by modern bacteria, and that any data generated is not authentically ancient, but instead from modern, contaminating bacterial or archael sequences. One way to distinguish between these two possibilities is to take advantage of the molecular clock, a concept that defines a near-constant rate of evolutionary change within organisms over time. Assuming a molecular clock, we would expect authentically ancient DNA sequences to contain many fewer mutations than contaminating, modern sequences, when compared to other modern data. We can therefore verify the authenticity of ancient sequences by asking whether they behave as expected for ancient sequences (fewer mutations along external branches in a phylogeny than modern sequences), or simply fall within the distribution of rates of the modern data included in the test. Using simulated and real bacterial gene sequence alignments, we developed a bioinformatics approach to evaluating the distribution of evolutionary rates along external branches in a phylgeny, which we have implemented in the popular software package BEAST. This method will be used to evaluate the results of the sequencing data recovered from both the Illumina sequencing and the Sanger sequencing of the cultured bacterial colonies.
Methane-derived isotopic & mineralogical biosignatures in cold seeps Orphan, House, Ferry, Freeman, and McKeegan (in collaboration with J. Grotzinger)
Other team members: Jeffrey Marlow (graduate student), Joshua Steele (postdoc), Anne Dekas (graduate student), Elizabeth Reichert (graduate student), David Case (graduate student), Grayson Chadwick (undergraduate).
This project is focused on the application of molecular and isotopic methods for the characterization of biosignatures associated with methane cycling in marine methane vents and associated authigenic carbonates. Recent progress in the area of biosignature characterization by members of the Caltech team have included the application of δD analysis of bacterial lipids recovered from diverse methane impacted marine sediments, isotopic analysis of F430 from methane seep sediments as a proxy for active archaeal methanotrophs and/or methanogens (in collaboration with K. Freeman at PSU), and the characterization of viable endolithic methanotrophs associated with deep-sea authigenic carbonates.
Active endolithic microorganisms and methane cycling in authigenic carbonates
Methane derived authigenic carbonates have been previously shown to harbor lipid and DNA biosignatures affiliated with methanotrophic archaea and sulfate-reducing bacteria. While these biosignatures are often interpreted as ‘fossil’ remnants of microorganisms in the literature, it is also possible that they are derived from viable endolithic microorganisms. Using FISH combined with assays of methanotrophic activity (14CH4 and CH3D), we documented abundant methanotrophic archaea/ SRB consortia within the interiors of authigenic carbonates from a variety of deep-sea habitats (at the seabed within active seeps and inactive areas as well as sediment hosted carbonate nodules). The aggregate morphology and abundance of these organisms varied according to habitat, with the largest archaeal biomass recorded in seafloor carbonates and nodules in proximity to active CH4 venting and approximately half as many cells in carbonates from inactive areas. In support of these observations, methane oxidation rates in these samples (assessed by the biological conversion of CH3D to D2O) followed a similar trend, with the highest rates of methanotrophy affiliated with active carbonates and nodules relative to carbonates from dormant seep areas. Stable isotope incubation experiments with 15NH4 and CH4 coupled with FISH-nanoSIMS analysis were also used to demonstrate active growth of methanotrophic consortia within the interiors of carbonate nodules (Marlow et al. In Review).
D/H analysis of bacterial lipids: constraining potential carbon metabolism by syntrophic sulfate-reducing bacteria
Over the past year, we have been collaborating with Alex Sessions (Caltech) to explore the use of D/H values of lipid biomarkers as an independent proxy for carbon metabolism by sulfate-reducing bacteria. Using a combination of environmental samples and laboratory incubations. We are studying the potential carbon metabolism of the sulfate reducing bacterial (SRB) partner affiliated with anaerobic oxidation of methane (AOM). Arguments have been made based upon the δ13C value of SRB associated fatty acids that the bacterial partner is an autotroph assimilating the majority of its carbon from seawater DIC (Wegener et al., 2008), while others suggest that the SRB partners are heterotrophs or mixotrophs deriving some of their carbon from compounds produced by methanotrophic archaea (Elvert et al., 2003; Neretin et al., 2007). To further elucidate the nature of the syntrophic relationship we are analyzing the hydrogen isotopic values of bacterial fatty acids extracted from sediment cores associated with a variety of active seep settings.
Previous work by Zhang et al (2009) identified patterns in the hydrogen isotopic fractionation of bacterial fatty acids based upon the central metabolic pathway. Hydrogen precursors for lipid synthesis include NADH, water and organic substrates. NADH is the primary biosynthetic precursor of hydrogen for lipids, representing about 50% of lipid hydrogen. Fractionation of NADH hydrogen is associated with its generation through different metabolic pathways, which can be observed through the growth of bacteria on different substrates (Zhang et al., 2009).
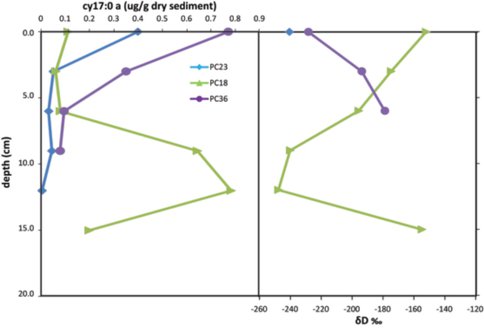
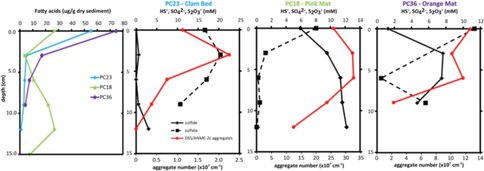
Environmental samples, including those analyzed from Hydrate Ridge (Figure 1), display greater than a 100‰ range in D/H content indicating contributions from multiple metabolic pathways and multiple microorganisms (Valentine, 2009). Specific fatty acids have been proposed as genera specific biomarkers for sulfate-reducing Deltaproteobacteria. These include a cyclopropyl-17:0 fatty acid (cy-17:0) in Desulfosarcina/ Desulfococcus (Elvert et al., 2003), the major bacterial group associated with ANME-2 aggregates (Schreiber et al., 2010) . Results from three locations at Hydrate Ridge show the greatest depletion in cy-17:0 at the depths where it is most abundant (Figures 1-2). However, the depths at which cy-17:0 is most concentrated do not correlate to the peaks in aggregate concentrations (Figure 2). With additional analysis of fatty acids from environmental samples, as well as cultures of SRB additional fatty acids may prove useful as biomarkers for the SRB of AOM aggregates. Additionally, D/H ratios of archaeal lipids from these samples and cultures may provide insight into intermediates associated with the syntrophic associations.
Figure 1: (left) Concentration of cyclopropyl 17:0 fatty acid, a putative sulfate reducing bacterial biomarker, with depth in three locations at Hydrate Ridge North. (right) δD values for cyclopropyl 17:0 fatty acid with depth show the most depleted values at the depths with the greatest concentration.
Figure 2: Depth profiles of fatty acid concentrations and pore water sulfide (solid-line, diamonds), sulfate (dashed-line, squares), and aggregate numbers (red) within cores from Hydrate Ridge North.
This research is fundamentally multidisciplinary in nature, requiring expertise in molecular biology, analytical microscopy, and organic and isotopic geochemistry. The combination of microbiological and geochemical methodologies is an effective strategy for connecting specific groups of microorganisms with their physiological potential and biosignatures. Our collaboration with Kate Freeman’s group focused on the development of isotopic analyses of coenzyme F430 in marine sediments is a direct outcome of the NAI and PSARC team.
Peer reviewed publications:
Marlow et al. (in review) Carbonate Hosted Methanotrophy: An Unrecognized Methane Sink in the Deep Sea. Nature Geosciences.
Abstracts presented:
DAWSON, K.S., SESSIONS, A.L. and ORPHAN, V.J. (2012) Using D/H fractionation in lipids to elucidate the metabolic role of sulfate reducing bacteria in AOM. [poster] _Gordon Research Symposium: Organic Geochemistry_, July 2012.
Life in Greenland glacial ice Brenchley (PI), Miteva (Sc Res Assoc), and Kaitlyn Rinehold (technician) with undergraduate students L. Seth, N. Lazar and C. Burlingame (in collaboration with Todd Sowers)
Last year, we developed and published a new rapid and sensitive method for assessing the relatedness of bacterial species (Loveland-Curtze J., Miteva V., Brenchley J. (2011), and performed microbiological and molecular analyses of decontaminated ice core samples from the recent NEEM ice core drill in Greenland. This year, improving methodological approaches for extracting DNA from ice core samples with low abundance of microbial cells and performing reliable diversity analyses was a major goal of our research. We evaluated and optimized a new DNA extraction procedure using Sterivex (Millipore) filtration for cell concentration instead of centrifugation followed by MoBio Ultraclean DNA extraction procedure with bead-beating. Results showed 5 to 10 times better yields when cell pellets were collected on Sterivex filters. However, because the genomic DNA yields from some samples are still low, we also use whole genome amplification with RepliG (Qiagen). Comparisons of parallel small subunit rRNA gene clone libraries from original and RepliG amplified DNA from the same Greenland ice core sample showed similar microbial diversity, represented by the same genera.
We focused our studies on comparisons of isolates obtained from four decontaminated samples from different depth, age and deposition climate of the NEEM Greenland ice core. Samples originated from depths 100m, 633m, 1731m and 2051m corresponding to 340, 3,220, 36,500 and 78,500 years before present. Ice from the first two samples was deposited during the present warmer interglacial period, while the deeper 1731 m sample contained ice deposited during a cold period. The highest cell abundance in the range of 1-5×106/ml was detected in the 100m and 1730m depth samples, followed by the deepest 2051m sample (4.6×105/ml) and the lowest cell counts were found at 633m (8.3×104/ml). Interestingly, after applying the same plating and cultivation approach, the highest number of isolates was obtained from the latter ice core sample of low cell abundance, bringing the level of culturability up to 0.05%, whereas in the other three samples it was at least 2 orders of magnitude lower. Diversity in the 633m ice sample was also high with isolates representing Proteobacteria, Firmicutes, Actinobacteria and fungi. Among these isolates was a novel microorganism affiliated with Alpha Proteobacteria based on 16S rRNA gene sequencing, with the closest validated relative Hansshlegelia plantiphila (a methanol utilizing bacterium) at a distance of 0.907. Further genetic and physiological analyses are aimed at the validation of this new bacterium as a possible new genus (or family).
Biosignatures of life in Archean-analog anoxic biofilms Macalady & Ph.D. students McCauley, Jones (in collaboration with A. Montanari, K. Broad, Italian cave explorers associated with the Federazione Speleologica Marchigiana)
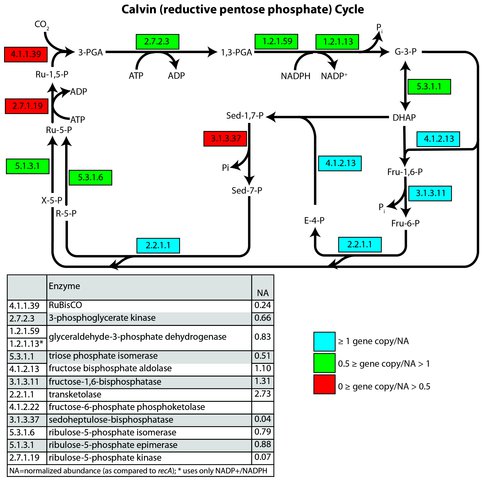
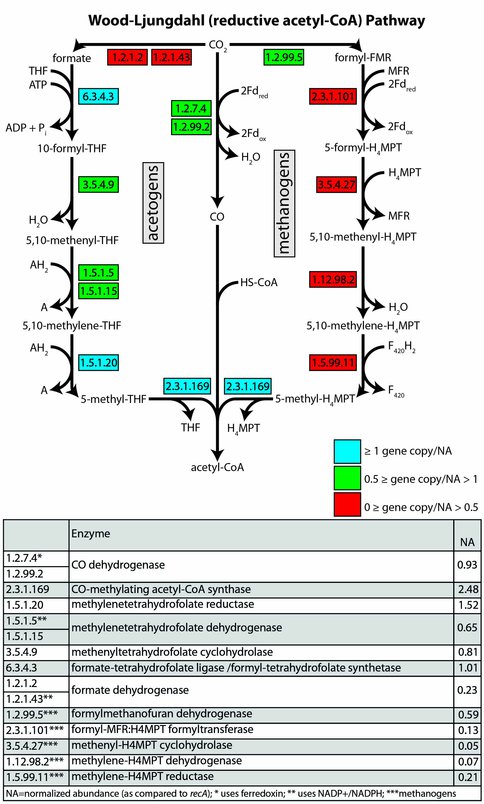
Astrobiology Dual Title graduate student Rebecca McCauley is characterizing microbial life in anoxic cave lakes ~600 m below ground surface. The groundwater hosting biofilms is being studied as an Archean analog using genetic and geochemical approaches. Ongoing metagenomic analyses from two environmental metagenomes completed earlier this year at the DOE Joint Genome Institute show that genes from multiple pathways for autotrophic metabolism (carbon fixation) are present. These results are significant to biosignatures because the isotopic signatures imparted by carbon fixation pathways differ, and can be preserved in ancient organic matter. The Calvin Cycle is present at low abundance in the community, but the Wood-Ljungdahl pathway is present at higher gene copy number (Figures 3 and 4). A publication is in preparation.
This project requires the cooperation and coordination of cave explorers (Broad, Italian cave explorers), geologists (Montanari) and microbial geochemists (Macalady, McCauley). Microbial geochemistry research itself is interdisciplinary in nature. For example, McCauley’s dissertation research encompasses both aqueous geochemistry (field and laboratory analyses, thermodynamic calculations) and metagenomic DNA sequence analysis using bioinformatics.
Figure 3: Gene copy number normalized by recA for the Lago Infinito biofilm community. Key enzymes are not highly abundant in the metagenome suggesting a minimal role of the Calvin Cycle in this system.
Figure 4: Gene copy number normalized by recA for the Lago Infinito biofilm community. Key enzymes are almost non-existent in the metagenome for methanogens, but of medium abundance for acetogens, suggesting the Wood-Ljungdahl pathway plays a sub-dominant role in this system.
Abstracts presented:
Macalady, J.L.*, D.S. Jones, B. Dempsey, L. Polerecky. 2012. Fate of sulfide in limestone aquifers and implications for cave formation. Astrobiology Science Conference (AbSciCon), Atlanta, GA, April 2012.
Macalady, J.L.* [invited]. 2012. Metagenome-enabled investigation of microbial sulfur precipitation in a carbonate aquifer. Goldschmidt Conference, Montreal, June 2012
Jones, D. S.*, Polerecky, L. Dempsey, B. A., Galdenzi, S. and Macalady, J.L. 2011. Biological and abiotic controls on sulfide oxidation, acid production, and carbonate dissolution in the Frasassi cave system, Italy GSA Annual Meeting, Oct. 11, 2011, Minneapolis, MN.
Macalady, J.L.* [keynote]. 2011. Direct human exploration of the terrestrial subsurface. Pardee Symposium on Exploration of the Deep Biosphere, GSA Annual Meeting, Oct. 11, 2011, Minneapolis, MN.
Macalady, J.L.* [keynote] 2011. Sulfidic caves as model systems for microbial biogeochemistry. International Society for Environmental Biogeochemistry, Istanbul, Turkey, Sept. 2011.
McCauley, R. L., D.S. Jones, I. Schaperdoth, L. Steinberg, J.L. Macalady. 2011. Metabolic Strategies in Energy-Limited Microbial Communities in the Anoxic Subsurface, Dark Energy Biosphere Initiative (DEBI) Meeting, Chapel Hill, NC. Poster.
Zerkle, A. L., Macalady, J. L., Jones, D. S., and Farquhar, J. (2012) S isotope investigation of sulfur cycling in the sulfidic Frasassi cave system: A case study of chemotrophic sulfide oxidation in the environment: EMBO Workshop on Microbial Sulfur Metabolism, Noordwijkerhout, The Netherlands
Biosignatures of life in ancient stratified ocean analogs Macalady, Freeman, & Kump with T. Hamilton (postdoc) and Ph.D. students J. Fulton, K. Meyer & R. McCauley (in collaboration with P. Welander (MIT), R. Summons (MIT), D. Newman (Caltech), Brian Kakuk (Bahamas Caves Research Foundation))
Instigated by Macalady and Kump in 2010, this project investigates biosignatures of life in modern analogs for stratified ancient and/or extraterrestrial oceans. A field workshop at meromictic Fayetteville Green Lake sponsored by NAI, Agouron Institute, and CIFAR was attended by 50 national and international participants last year. Emerging from the workshop, a special issue of Geobiology devoted to biosignatures in anaerobic photic ecosystems has been published, edited by Macalady and colleagues Julia Maresca (U. Delaware) and Sean Crowe (U. of Southern Denmark).
A field expedition to stratified Bahamian blue hole Magical Sink was led by Astrobiology dual-title Ph.D. student Rebecca McCauley in 2011. Measurements of photosynthetically active radiation (PAR) collected in collaboration with technical diver and cave explorer Brian Kakuk confirmed that anoxygenic phototrophs in the sinkhole grow using light levels similar to or lower than those at the bottom of the Black Sea chemocline, thus making the microbial species involved candidates for the earth’s most extreme low-light phototrophs. A manuscript is in preparation for publication.
A new Proterozoic-analog microbial ecosystem was discovered last year and has unique potential to help us understand biosignatures of earth evolution, specifically the long delay in the continued oxidation of the surface earth after the initial rise at the Great Oxidation Event (GOE, ~2.4 Ga). Based on initial findings, Little Salt Spring, Florida is weakly stratified and has low concentrations of oxygen, sulfide and sulfate throughout the water column, similar to the geochemistry hypothesized for Proterozoic oceans. The sinkhole hosts a mixed oxygenic/anoxygenic community of microbial phototrophs. This site thus provides an opportunity to investigate biogeochemical and microbial processes in a Proterozoic ocean analog, including the ecological and geochemical controls on oxygen production and organic matter preservation. In addition, the microbial ecosystem at the site produces large quantitites of hopanoids, an important class of organic biomarkers that can be preserved in rocks for billions of years. Freeman lab Ph.D. student Laurence Bird participated in a site reconnaissance and sampling expedition in summer 2012 along with PSARC postdoc Trinity Hamilton. Bird collected sediment and biofilm samples along the water column redox gradient for organic biomarker analyses including hopanoids. A sample from the site was analyzed by Paula Welander and Dianne Newman’s lab, confirming cyanobacterial production of the important biomarker 2-methyl hopanoid in the biofilm. The MIT and Caltech collaborations are a direct result of NAI interactions.
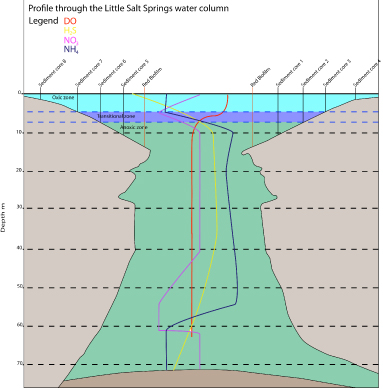
Figure 5. Geochemistry of the Little Salt Spring sinkhole in June 2012.
A website monitoring the activities of an informal working group on Early Earth Photosynthesis is maintained by Macalady (http://www.geosc.psu.edu/~jlm80/EEP.html).
Peer reviewed publications:
Maresca, J., Crowe, S. and Macalady, J. L. 2012. Guest Editorial: Special Issue on Anaerobic Photosynthetic Ecosystems. Geobiology 10 (3): 193-195.
Abstracts presented:
Meyer, K. M., Fulton, J. M.*, Hunter, S., Macalady, J. L., Kump. L., and Freeman, K.H. [invited]. 2012. Dynamics of the anoxygenic phototrophic community in meromictic Fayetteville Green Lake (NY) and the associated sedimentary pigment record. American Geophysical Union Annual Meeting, December
Macalady, J.L.* [invited]. 2012. Energy, ecology, and the distribution of microbial life. Royal Society Discussion Meeting on Energy Transduction and Genome Function, London, England, November 11-12.
Macalady, J.L.* [invited] 2012. Microbial authors of earth’s ancient record: which microbes left their mark? Kavli Frontiers of Science Symposium, Newport Beach, CA, Nov. 1-4.
Macalady, J.L.*, R. L. McCauley, I. Schaperdoth, B. Kakuk. 2012. Extremely low-light adapted phototrophic biofilm community in a Bahamian blue hole. Astrobiology Science Conference (AbSciCon), Atlanta, GA, April 2012.
Ca isotopic biosignatures project (new)
Faculty: Matthew Fantle (PSU), Jenn Macalady (PSU)
External NAI members: Marilyn Fogel (CIW)
External faculty: Anton Eisenhauer (GEOMAR)
Staff: Irene Shaperdoth (PSU); Ana Kolevica (GEOMAR)
Students: Khadouja Harouaka, PhD student, Geosciences (PSU); Matthew Gonzales, PhD student, Geosciences (PSU); Amanda Fobes, Geosciences undergraduate (PSU); Muammar Mansor, Geosciences undergraduate.
The overall goal of the research is to constrain the potential for microbes to affect the Ca isotopic composition of minerals in nature. We are testing this experimentally by characterizing abiotic Ca isotope fractionation during gypsum precipitation as a function of precipitation rate, saturation state, and Ca:SO4 ratio. Natural samples of gypsum are then collected and measured for Ca and S isotopes, particle size, and particle morphology to determine if Ca isotopes might serve as a biosignature in the environment. This work will lead to a greater fundamental understanding of the controls on Ca isotopic fractionation in nature, which enables an evaluation of its general utility as a biosignature.
Two primary research goals were advanced this year: (1) measurement of natural gypsum samples from the sulfidic Frasassi cave system, and
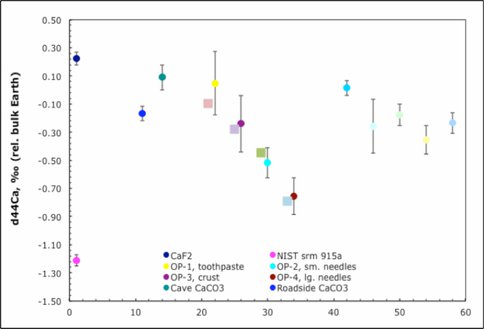
Figure 6. Ca isotope measurements of samples from the Frasassi cave system, Italy and surroundings.
(2) Ca and S isotope measurements of experimentally-precipitated gypsum samples.
Measurements of experimental products also included particle size distributions, BET surface area measurements, and SEM imaging.
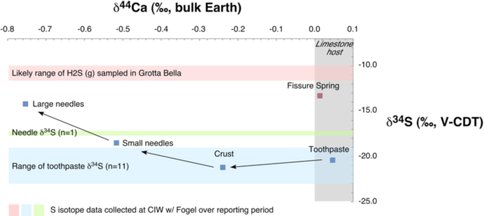
Figure 7. Ca and S isotopic composition of gypsum and H2S (g) samples from the Frasassi cave system, Italy.
This project brings together an isotope geochemist (Fantle) and a microbial geochemist (Macalady). The collaboration extends to the students involved in the project. Harouaka (Geosciences) collaborated with Mansor, an undergraduate researcher (B.S. Biotechnology) to precipitate gypsum in the presence of Acidithiobacillus thiooxidans.
This project spawned collaborations between Fantle and European colleagues (Eisenhauer, GEOMAR, Kiel, Germany) and colleagues at the Carnegie Institution of Washington (Fogel). The presence of NAI led directly to these specific collaborations, and the NAI link between PSU and CIW definitely facilitated the S isotope work.
Publications:
Harouaka, K. 2011. An investigation of the mechanisms of calcium isotopic fractionation in gypsum. M.S. thesis.
Publications
-
Dawson, K. S., Freeman, K. H., & MacAlady, J. L. (2012). Molecular characterization of core lipids from halophilic archaea grown under different salinity conditions. Organic Geochemistry, 48, 1–8. doi:10.1016/j.orggeochem.2012.04.003
-
Maresca, J. A., Crowe, S. A., & MacAlady, J. L. (2012). Anaerobic photosynthetic ecosystems. Geobiology, 10(3), 193–195. doi:10.1111/j.1472-4669.2012.00327.x
-
Rhodes, M. E., Oren, A., & House, C. H. (2012). Dynamics and Persistence of Dead Sea Microbial Populations as Shown by High-Throughput Sequencing of rRNA. Applied and Environmental Microbiology, 78(7), 2489–2492. doi:10.1128/aem.06393-11
-
Schopf, J. W., Farmer, J. D., Foster, I. S., Kudryavtsev, A. B., Gallardo, V. A., & Espinoza, C. (2012). Gypsum-Permineralized Microfossils and Their Relevance to the Search for Life on Mars. Astrobiology, 12(7), 619–633. doi:10.1089/ast.2012.0827
- Marlow et al. (2012, In Review). Carbonate Hosted Methanotrophy: An Unrecognized Methane Sink in the Deep Sea. Nature Geosciences.
- Nasdala, L., Beyssac, O., Schopf, J.W. & Bleisteiner, B. (2012). Application of Raman-based images in the Earth sciences. In: Zoubir, A. (Eds.). Raman Imaging, Springer Series in Optical Sciences. Vol. 168. Amsterdam: Springer.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
James Ferry
Co-Investigator
Katherine Freeman
Co-Investigator
James Schopf
Co-Investigator
Beth Shapiro
Co-Investigator
Oded Beja
Collaborator
David Bice
Collaborator
Kenneth Broad
Collaborator
Charles Cockell
Collaborator
Anton Eisenhauer
Collaborator
Jack Farmer
Collaborator
Sorel Fitz-Gibbon
Collaborator
Marilyn Fogel
Collaborator
Brian Kakuk
Collaborator
Anatoliy Kudryavtsev
Collaborator
Kevin McKeegan
Collaborator
Alessandro Montanari
Collaborator
Aharon Oren
Collaborator
John Spear
Collaborator
Roger Summons
Collaborator
John Valley
Collaborator
Martin Van Kranendonk
Collaborator
Mary Voytek
Collaborator
Malcolm Walter
Collaborator
Paula Welander
Collaborator
Aubrey Zerkle
Collaborator
Jake Bailey
Postdoc
Trinity Hamilton
Postdoc
Theresa M. D. Raub
Postdoc
Tim Raub
Postdoc
Joshua Steele
Postdoc
Lisa Steinberg
Postdoc
Jennifer Loveland-Curtze
Research Staff
Vanya Miteva
Research Staff
Irene Schaperdoth
Research Staff
Zhidan Zhang
Research Staff
Heidi Albrecht
Doctoral Student
Laurence Bird
Doctoral Student
David Case
Doctoral Student
Khadouja Harouaka
Doctoral Student
Daniel Jones
Doctoral Student
Kristine Korzow Richter
Doctoral Student
Muammar Mansor
Doctoral Student
Jeffrey Marlow
Doctoral Student
Rebecca McCauley
Doctoral Student
Elizabeth Reichert
Doctoral Student
Moshe (Mathew) Rhodes
Doctoral Student
Abigail M. Green
Graduate Student
Benjamin Harrison
Graduate Student
Norbert Lazar
Graduate Student
Thomas Roberts
Graduate Student
Labdhi Seth
Graduate Student
Anne Dekas
Unspecified Role
Matthew Gonzalez
Unspecified Role
Reinhard Kozdon
Unspecified Role
A. Nele Meckler
Unspecified Role
Mathias Stiller
Unspecified Role
Kenneth Williford
Unspecified Role
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.3
Effects of extraterrestrial events upon the biosphere
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems