2012 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Georgia Institute of Technology
Reporting | SEP 2011 – AUG 2012
Ironing Out the RNA World
Project Summary
In RNA World models of evolution, RNA was once the primary biopolymer of genetics and catalysis (1). Ancient RNA-based life would have inhabited an earth with abundant soluble iron and no free oxygen (2,3). Anoxic life persisted for around 1.0-1.5 billion years before photosynthesis began producing substantial free oxygen. The ‘great oxidation’ led to Fe2+/O2 mediated cellular damage (4) and depletion of soluble iron from the biosphere (5). We hypothesize that Fe22+ was an RNA cofactor when iron was benign and abundant and that Fe2+ was replaced by Mg2+ during the great oxidation. The RNA-Fe2+ to RNA-Mg2+ hypothesis is in close analogy with known metal substitutions in some metalloproteins (6-11). An ancestral ribonucleotide reductase (RNR), for example, spawned di-iron, di-manganese, and iron-manganese RNRs (12). Our hypothesis is supported by observations (13) that (i) RNA folding is conserved between complexes with Fe2+ and Mg2+ and (ii) at least some phosphoryl transfer ribozymes are more active in the presence of Fe2+ than Mg2+. Here, we demonstrate that reversing the putative metal substitution in an anoxic environment, by removing Mg2+ and adding Fe2+, expands the catalytic repertoire of some RNAs. Fe2+ can confer on RNA a previously uncharacterized ability to catalyze single electron transfer. Catalysis is specific, in that it is dependent on the type of RNA. The 23S rRNA and tRNA, some of the most abundant and ancient RNAs (14), are found to be efficient electron transfer ribozymes in the presence of Fe2+. Therefore, the catalytic competence of ancient RNAs may have been greater in early earth conditions than in extant conditions, and the experiments described here may be reviving latent function.
Project Progress
Without cations, RNA is essentially incapable of molecular recognition or catalysis. RNA requires cations in the form of Na+, K+ and Mg2+ for folding and function (15-17). Mg2+ was originally demonstrated to be especially important in folding of tRNA (18-20) and is now known to be critical for folding of compact RNAs. Mg2+ ions neutralize the negative charge of the RNA backbone and bind specifically to complex structural features of RNA (21). Mg2+ is required by many ribozymes for organizing RNA or water molecules within active sites, and for stabilizing reactants or transition states (22,23).
Using a standard peroxidase assay (24), we investigated the catalytic abilities of a variety of native nucleic acids to determine if they can catalyze single electron transfer in association with Fe2+ instead of Mg2+ . In this assay, an organic dye (a reducing agent) is oxidized by hydrogen peroxide (an oxidizing agent) via electron transfer. Specifically, one electron is transferred from 3,3′,5,5′-tetramethylbenzidine (TMB) to hydrogen peroxide, forming a radical cation (TMB• +) according to the equation:
½ (HOOH) + TMB + H+ → H2O + TMB• +.
This reaction is a broadly used assay for detection of electron transfer. Absorbance at 370 nm and 652 nm is used to monitor the course of the reaction.
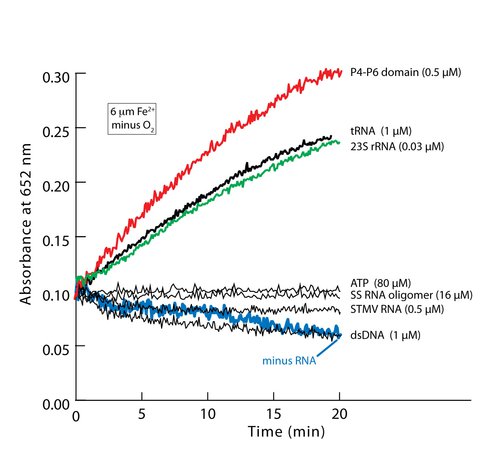
Catalysis of single electron transfer is observed for a subset of RNAs tested here (Figure 1). Fe2+ , RNA, and the exclusion of O2 are required. In solutions containing Mg2+ instead of Fe2+ , solutions lacking RNA, or solutions with heat-degraded RNA, catalysis of electron transfer is not detected. Some RNAs catalyze electron transfer much more efficiently than other RNAs, at lower concentration of RNA (0.03-1.0 μM strand, 75-100 μM in nucleotide). For the inefficient RNAs, electron transfer is observable only at high concentration (>500 μM in nucleotide). Relative rates of electron transfer are inferred from the initial slopes of graphs such as those represented in Figure 1. Additional information on materials, methods, reaction rates, RNA stability, and control reactions is found in the online-only methods.
The 23S rRNA gives efficient catalysis of electron transfer at a low concentration of RNA strands (Figure 1), with a greater kcat (Figure 2) than other RNAs. On a per nucleotide bases, the P4-P6 domain RNA and a mixture of yeast tRNAs are most efficient, catalyzing electron transfer with roughly similar efficiencies. The appropriate choice of concentration units, strand or nucleotide, is dependent on whether the Fe2+ binds at a few highly specific sites, or less specifically to many sites on the RNAs.
We examined a diverse set of nucleic acids in an attempt to isolate properties responsible for catalysis of electron transfer in the presence of Fe2+ (Figure 1). Closely spaced phosphate groups, as in ATP, are not sufficient to induce catalysis. The regular array of phosphate groups and other structural features of B-form DNA are insufficient for efficient catalysis. We investigated a double-stranded 32 base pair DNA duplex composed of a mixture of C-G and A-T base pairs. The combination of B-DNA and Fe2+ does not catalyze electron transfer. To investigate whether a short unstructured RNA catalyzes electron transfer in the presence of Fe2+ , we used a small, single-stranded RNA oligomer (5′-GCACU-3′). This short RNA oligomer does not catalyze electron transfer. We investigated the Satellite Tobacco Mosaic Virus (STMV) genomic RNA (25) to determine if a large, partially unstructured RNA is competent to catalyze electron transfer in the presence of Fe2+ . This 1058 nucleotide RNA genome does not catalyze electron transfer at 0.5 μM strand, which, in terms of nucleotide concentration, greatly exceeds the concentration where P4-P6 Domain RNA, 23S rRNA and tRNAs are catalytic.
The differential catalysis of single-electron transfer by various RNAs supports a model in which the potential to form well-defined three-dimensional RNA structure is important to catalysis. The ability of RNA to coordinate divalent cations may be significant. In the large ribosomal subunit a subset of associated divalent cations are chelated by up to three phosphate groups of the 23S rRNA (26,27). The P4-P6 domain RNA chelates fewer divalent cations but with analogous coordination geometry (28). tRNA also interacts strongly with divalent cations (18-20). In contrast, RNAs such as nucleotides, small single-stranded RNA, B-DNA, and the STMV RNA genome, which lack the potential to form well-defined structures and do not chelate divalent cations, are much less efficient electron transfer catalysts.
Fe2+ can substitute for Mg2+ in RNA compaction, as previously demonstrated for the P4-P6 domain RNA, which folds to the same state with either Mg2+ or Fe2+ ions (13), with common binding sites for either metal (29). However, the low salt conditions that yield the best catalysis here suggest that the RNAs are not fully folded to their native states in our experiments and therefore, the structures of RNA-Fe2+ complexes that yield catalysis remain to be fully characterized.
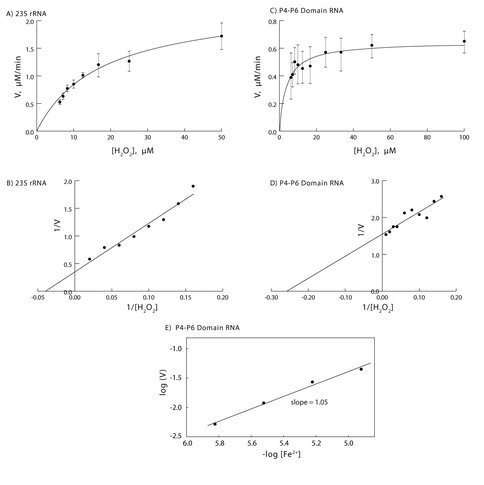
The catalytic nature of these electron transfer reactions is supported by standard enzyme assays. Since the amount of enzyme in a reaction mixture can be limiting, enzyme-catalyzed reaction rates plateau rather than increase monotonically with increasing substrate concentration. For both 23S rRNA and the P4-P6 Domain RNA, the electron transfer reaction does indeed saturate (Figures 2A & 2C).
Kinetic parameters for single electron transfer by 23S rRNA and the P4-P6 Domain RNA with Fe2+ were determined by fitting data to a Michaelis-Menten model (Figure 2). Non-linear regression analysis of the experimental data shows that for P4-P6 domain RNA, kcat = 2.2 10-2/s, Km = 4.15 10-6 M, kcat /Km = 5.3 103/M⋅s. For the 23S rRNA, kcat =1.28/s, Km = 1.75 10-5 M, kcat /Km = 7.3 104/M⋅s. However, if the RNA is not saturated with Fe2+ , the data underestimate the true kcat.
Previously Breaker isolated a series of hammerhead ribozymes that cleave RNA upon binding to divalent metal cations (30). Those phosphoryl-transfer ribozymes show linear relationship of log rate versus log [cation] for RNA cleavage. Slopes of either one or two in the log-log graphs were interpreted as indicating either one or two cations being required for catalysis. Using Breaker’s approach here, for the P4-P6 Domain RNA, the log of the rate varies linearly with the log [Fe2+ ] (Figure 2E). The slope is approximately one. Although more complex models cannot be excluded, these observations are consistent with a mechanism in which occupancy of a single Fe2+ site confers catalytic activity to the RNA.
We observe that replacement of Mg2+ by Fe2+ alters and expands the functional capabilities of some biological RNAs to include redox-activity. Previously Suga used in vitro selection to obtain redox-activity of RNA. Suga’s activity requires covalent attachment of substrate to RNA (31). Sen showed that RNA can enhance the redox-activity of iron-protoporphyrin IX (32). By contrast, we observe that some of the most abundant and evolutionarily conserved RNAs have intrinsic redox functionality that is activated simply by interaction with Fe2+ . Our results here, combined with previous observations of binding of Fe2+ to RNA in vivo (33,34), suggest a biological collaboration of iron and RNA. We propose that RNA function, in analogy with protein function, can be fully understood only in the context of association with a range of possible metals. Our results add a new dimension to the RNA World hypothesis. RNA sequence space is clearly more densely occupied, with a broader array of function, than previously expected. This expansion of the apparent catalytic power of RNA suggests that sophisticated biochemical transformations, such as reduction of ribonucleotides to deoxyribonucleotides, were possible in an RNA world.
References 1. In Atkins, J. F., Gesteland, R. F. and Cech, T. R. (eds.). Cold Spring Harbor Laboratory Press.2. Anbar, A.D. (2008) Oceans. Elements and Evolution. Science (New York, N.Y.), 322, 1481-1483.
3. Hazen, R.M. and Ferry, J.M. (2010) Mineral Evolution: Mineralogy in the Fourth Dimension. Elements, 6, 9-12.
4. Prousek, J. (2007) Fenton Chemistry in Biology and Medicine. Pure Appl. Chem., 79, 2325-2338.
5. Klein, C. (2005) Some Precambrian Banded Iron-Formations (BIFs) from around the World: Their Age, Geologic Setting, Mineralogy, Metamorphism, Geochemistry, and Origin. Am. Mineral., 90, 1473-1499.
6. Aguirre, J.D. and Culotta, V.C. (2012) Battles with Iron: Manganese in Oxidative Stress Protection. J. Biol. Chem., 287, 13541-13548.
7. Ushizaka, S., Kuma, K. and Suzuki, K. (2011) Effects of Mn and Fe on Growth of a Coastal Marine Diatom Talassiosira Weissflogii in the Presence of Precipitated Fe(III) Hydroxide and EDTA-Fe(III) Complex. Fish. Sci., 77, 411-424.
8. Martin, J.E. and Imlay, J.A. (2011) The Alternative Aerobic Ribonucleotide Reductase of Escherichia Coli, Nrdef, Is a Manganese-Dependent Enzyme That Enables Cell Replication During Periods of Iron Starvation. Mol. Microbiol., 80, 319-334.
9. Cotruvo, J.A. and Stubbe, J. (2011) Class I Ribonucleotide Reductases: Metallocofactor Assembly and Repair in Vitro and in Vivo. Annu. Rev. Biochem, 80, 733-767.
10. Anjem, A., Varghese, S. and Imlay, J.A. (2009) Manganese Import Is a Key Element of the Oxyr Response to Hydrogen Peroxide in Escherichia Coli. Mol. Microbiol., 72, 844-858.
11. Wolfe-Simon, F., Starovoytov, V., Reinfelder, J.R., Schofield, O. and Falkowski, P.G. (2006) Localization and Role of Manganese Superoxide Dismutase in a Marine Diatom. Plant Physiol., 142, 1701-1709.
12. Torrents, E., Aloy, P., Gibert, I. and Rodriguez-Trelles, F. (2002) Ribonucleotide Reductases: Divergent Evolution of an Ancient Enzyme._ J. Mol. Evol._, 55, 138-152.
13. Athavale, S.S., Petrov, A.S., Hsiao, C., Watkins, D., Prickett, C.D., Gossett, J.J., Lie, L., Bowman, J.C., O’Neill, E., Bernier, C.R. et al. (2012) RNA Folding and Catalysis Mediated by Iron (II). PLoS ONE, 7, e38024.
14. Fox, G.E. (2010) Origin and Evolution of the Ribosome. Cold Spring Harb. Perspect. Biol., 2, a003483.
15. Bowman, J.C., Lenz, T.K., Hud, N.V. and Williams, L.D. (2012) Cations in Charge: Magnesium Ions in RNA Folding and Catalysis Curr. Opin. Struct. Biol., 22, 262-272.
16. Auffinger, P., Grover, N. and Westhof, E. (2011) Metal Ion Binding to RNA. Met Ions Life Sci, 9, 1-35.
17. Brion, P. and Westhof, E. (1997) Hierarchy and Dynamics of RNA Folding. Annu. Rev. Biophys. Biomol. Struct., 26, 113-137.
18. Stein, A. and Crothers, D.M. (1976) Conformational Changes of Transfer RNA. The Role of Magnesium(II). Biochemistry, 15, 160-168.
19. Lynch, D.C. and Schimmel, P.R. (1974) Cooperative Binding of Magnesium to Transfer Ribonucleic Acid Studied by a Fluorescent Probe. Biochemistry, 13, 1841-1852.
20. Lindahl, T., Adams, A. and Fresco, J.R. (1966) Renaturation of Transfer Ribonucleic Acids through Site Binding of Magnesium. Proc. Natl. Acad. Sci. U. S. A., 55, 941-948.
21. Petrov, A.S., Bernier, C.R., Hsiao, C.L., Okafor, C.D., Tannenbaum, E., Stern, J., Gaucher, E., Schneider, D., Hud, N.V., Harvey, S.C. et al. (2012) RNA-Magnesium-Protein Interactions in Large Ribosomal Subunit. J. Phys. Chem. B, 116, 8113-8120.
22. Butcher, S.E. (2011) The Spliceosome and Its Metal Ions. Met Ions Life Sci, 9, 235-251.
23. Johnson-Buck, A.E., McDowell, S.E. and Walter, N.G. (2011) Metal Ions: Supporting Actors in the Playbook of Small Ribozymes. Met Ions Life Sci, 9, 175-196.
24. Josephy, P.D., Eling, T. and Mason, R.P. (1982) The Horseradish Peroxidase-Catalyzed Oxidation of 3,5,3’,5’-Tetramethylbenzidine. Free Radical and Charge-Transfer Complex Intermediates. J. Biol. Chem., 257, 3669-3675.
25. Larson, S.B., Day, J., Greenwood, A. and McPherson, A. (1998) Refined Structure of Satellite Tobacco Mosaic Virus at 1.8 Å Resolution. J. Mol. Biol., 277, 37-59.
26. Hsiao, C. and Williams, L.D. (2009) A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res., 37, 3134-3142.
27. Klein, D.J., Moore, P.B. and Steitz, T.A. (2004) The Contribution of Metal Ions to the Structural Stability of the Large Ribosomal Subunit. RNA, 10, 1366-1379.
28. Cate, J.H., Hanna, R.L. and Doudna, J.A. (1997) A Magnesium Ion Core at the Heart of a Ribozyme Domain. Nat. Struct. Biol., 4, 553-558.
29. Berens, C., Streicher, B., Schroeder, R. and Hillen, W. (1998) Visualizing Metal-Ion-Binding Sites in Group I Introns by Iron(II)-Mediated Fenton Reactions. Chem. Biol., 5, 163-175.
30. Zivarts, M., Liu, Y. and Breaker, R.R. (2005) Engineered Allosteric Ribozymes That Respond to Specific Divalent Metal Ions. Nucleic Acids Res., 33, 622-631.
31. Tsukiji, S., Pattnaik, S.B. and Suga, H. (2004) Reduction of an Aldehyde by a Nadh/Zn2+ -Dependent Redox Active Ribozyme. J. Am. Chem. Soc., 126, 5044-5045.
32. Sen, D. and Poon, L.C. (2011) RNA and DNA Complexes with Hemin [Fe(III) Heme] Are Efficient Peroxidases and Peroxygenases: How Do They Do It and What Does It Mean? Crit. Rev. Biochem. Mol. Biol., 46, 478-492.
33. Ma, J., Haldar, S., Khan, M.A., Sharma, S.D., Merrick, W.C., Theil, E.C. and Goss, D.J. (2012) Fe2+ Binds Iron Responsive Element-RNA, Selectively Changing Protein-Binding Affinities and Regulating mRNA Repression and Activation. Proc. Natl. Acad. Sci. U. S. A., 109, 8417-8422.
34. Serra, M.J., Baird, J.D., Dale, T., Fey, B.L., Retatagos, K. and Westhof, E. (2002) Effects of Magnesium Ions on the Stabilization of RNA Oligomers of Defined Structures. Rna-a Publication of the Rna Society, 8, 307-323.
Electron transfer reactions catalyzed by RNA-Fe2+. Some RNAs, in the presence of Fe2+, catalyze electron transfer. The 23S rRNA from T. thermophilus, the P4-P6 domain of the T. thermophila Group 1 intron, and tRNA are catalytic. Other nucleic acids including ATP, a short RNA oligomer, double-stranded DNA, and the genome of STMV are inefficient catalysts. All reactions were performed in the absence of O2 and Mg2+, and in the presence of Fe2+ and H2O2. Concentrations are given in units of molecules (single strands). The concentration per nucleotide is roughly the same for ATP, the SS RNA oligomer, P4-P6 domain RNA, 23S rRNA, tRNA and DNA (i.e., ca. 80 μM), and is greater for STMV genome (i.e., ca. 500 μM).
Kinetics of RNA/Fe2+ catalyzed electron transfer. A) The electron transfer reaction catalyzed by 23S rRNA of T. thermophilus is saturable, reaching a plateau at high [H2O2]. B) The kinetics of the 23S rRNA reaction can be fit to a Michaelis-Menten model. C) The electron transfer reaction catalyzed by the P4-P6 domain of the T. thermophila Group 1 intron is saturable, reaching a plateau at high [H2O2]. D) The kinetics of the P4-P6 domain RNA reaction can be fit to a Michaelis-Menten model. E) The Fe2+ dependence of the reaction catalyzed by P4-P6 domain RNA suggests one Fe2+ is involved in catalysis. The reactions in panels A & B were performed with 0.03 μM 23S rRNA, 6 μM Fe2+, pH 6.5, 500 μM TMB. The reactions in panels C & D were performed with 0.5 μM P4-P6 domain RNA, 6 μM Fe2+, pH 6.5, 500 μM TMB. The reactions in panel E were performed with 0.50 μM P4-P6 domain RNA, 50 μM H2O2, pH 6.5, 500 μM TMB. For all reactions, O2 was excluded. Concentrations are in units of strands. The lines in panels B, D and E were obtained by linear regression. The RNA concentration units are strand.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Shreyas Athavale
Postdoc
Chiaolong Hsiao
Postdoc
Anton Petrov
Postdoc
J. Derrick Watkins
Postdoc
Jessica Bowman
Research Staff
I-Chun Chou
Research Staff
Eric O'Neill
Research Staff
Caitlin Prickett
Research Staff
Chad Bernier
Graduate Student
Denise Enekwa
Graduate Student
Jared Gossett
Graduate Student
Lively Lie
Graduate Student
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.