2012 Annual Science Report
 NASA Ames Research Center
Reporting | SEP 2011 – AUG 2012
NASA Ames Research Center
Reporting | SEP 2011 – AUG 2012
Origins of Functional Proteins and the Early Evolution of Metabolism
Project Summary
The main goal of this project is to identify critical requirements for the emergence of biological complexity in early habitable environments by examining key steps in the origins and early evolution of functional proteins and metabolic reaction networks. In particular, we investigate whether protein functionality can arise from an inventory of polypeptides that might have naturally existed in habitable environments. We attempt the first demonstration of multiple origins of a single enzymatic function. We investigate experimentally how primordial proteins could evolve through the diversification of their structure and function and thus demonstrate key steps in the earliest evolution of protein functions.
Project Progress
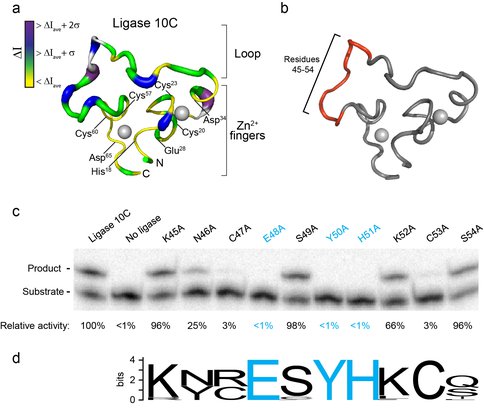
The structure and dynamics of a small enzyme capable of ligating two RNA fragments with the rate of 106 above background was fully characterized. This enzyme was evolved in vitro from a vast library of randomized proteins based on a scaffold. The enzyme does not resemble any contemporary protein. It consists of a flexible loop, a part of which is responsible for catalytic activity, a small, rigid core containing two zinc ions coordinated by neighboring amino acids and two highly flexible tails that might be of minor importance to the catalytic activity. In contrast to other zinc finger proteins this enzyme does not contain any ordered secondary structure elements such as helices or sheets. The ends of the loop are kept in direct proximity just through interactions of a charged residue and a histidine with a zinc ion, which they coordinate on the opposite side of the loop. Such a structure appears to be very fragile but, surprisingly, experiments and computer simulations of the original protein and its mutants indicate otherwise (see Fig. 1). The high flexibility of the protein facilitates its structural adjustments.
Figure 1. Probing the substrate-binding region of an artificial ligase enzyme. a) Thicker lines and darker colors in enzyme structure represent regions affected by enzyme-substrate complex formation measured as NMR chemical shift perturbations.
b) Region of ten single alanine mutants tested shown in red.
c) Ligase activity assay for alanine mutants by gel shift. The lower band corresponds to unligated substrate, and the upper band corresponds to ligated product. Activity values are normalized to the ligase free of mutations. Residues with activity below detection limit are shown in blue.
d) Sequence conservation analysis of previously identified ligase variants confirms the crucial importance of amino acids E, Y and H (blue).
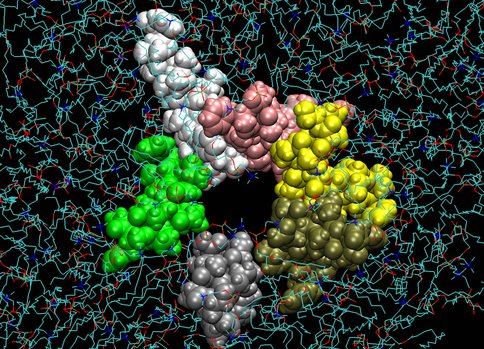
A similar picture emerges from studies of simple transmembrane channels that mimic those in ancestral cells. One such channel is an aggregation of an antiamoebin peptide that consists of only 16 amino acids. We found that this channel, in contrast to all known genomically coded, well-structured channels, is extremely flexible and does not form a conventional pore (Fig. 2). Yet it efficiently mediates ion transport.
Figure 2. Top view of the antiamoebin channel that is formed through aggregation of 6 monomers (yellow, gold, gray, green, white and pink) surrounding a water-filled pore embedded in the membrane. Water molecules were removed for clarity. This picture is a snapshot from molecular dynamics computer simulations. Note the highly irregular, asymmetric shape of the channel.
Taken together, these results indicate that highly flexible proteins or protein assemblies that do not resemble their contemporary counterparts could carry out functions quite efficiently. These might be points on a continuous evolutionary trajectory that form the “missing link” between simple, but only weakly active, oligopeptides and well-folded proteins similar to those found in modern organisms.
Publications
-
Chao, F-A., Morelli, A., Iii, J. C. H., Churchfield, L., Hagmann, L. N., Shi, L., … Seelig, B. (2012). Structure and dynamics of a primordial catalytic fold generated by in vitro evolution. Nat Chem Biol, 9(2), 81–83. doi:10.1038/nchembio.1138
-
Golynskiy, M. V., Haugner, J. C., Morelli, A., Morrone, D., & Seelig, B. (2013). In Vitro Evolution of Enzymes. Enzyme Engineering, None, 73–92. doi:10.1007/978-1-62703-293-3_6
-
Pohorille, A. (2012). Processes that Drove the Transition from Chemistry to Biology: Concepts and Evidence. Orig Life Evol Biosph, 42(5), 429–432. doi:10.1007/s11084-012-9304-3
-
Pohorille, A., & Pratt, L. R. (2012). Is Water the Universal Solvent for Life?. Orig Life Evol Biosph, 42(5), 405–409. doi:10.1007/s11084-012-9301-6
-
Wei, C., & Pohorille, A. (2013). Permeation of Aldopentoses and Nucleosides Through Fatty Acid and Phospholipid Membranes: Implications to the Origins of Life. Astrobiology, 13(2), 177–188. doi:10.1089/ast.2012.0901
- Pohorille, A. (2012, In Press). A few comments on Dynamic Kinetic Stability. Origins of Life and Evolution of the Biosphere.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Burckhard Seelig
Co-Investigator
Michael Wilson
Co-Investigator
James Lake
Collaborator
Chenyu Wei
Research Staff
Frank Chao
Graduate Student
Aleardo Morelli
Graduate Student
Thuy Nguyen
Graduate Student
Gareth Shannon
Graduate Student
Lewis Churchfield
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems