2011 Annual Science Report
 University of Hawaii, Manoa
Reporting | SEP 2010 – AUG 2011
University of Hawaii, Manoa
Reporting | SEP 2010 – AUG 2011
Amino Acid Alphabet Evolution
Project Summary
We study the question why did life on this planet “choose” a set of 20 standard building blocks (amino acids) for converting genetic instructions into living organisms? The evolutionary step has since been used to evolve organisms of such diversity and adaptability that modern biologists struggle to discover the limits to life-as-we-know-it. Yet the standard amino acid alphabet has remained more or less unchanged for 3 billion years.
During the past year, we have found that the sub-set of amino acids used by biology exhibits some surprisingly simple, strikingly non-random properties. We are now building on this finding to solidify a new insight into the emergence of life here, and what it can reveal about the distribution and characteristics of life elsewhere in the universe.
Project Progress
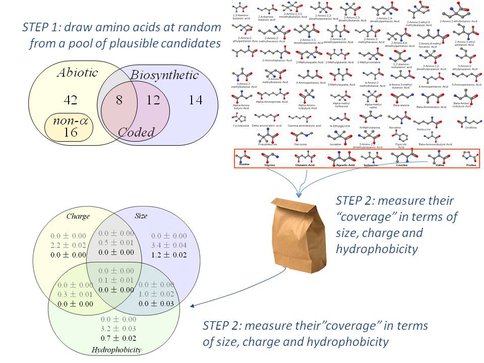
In 2010 we began analyzing the near-infinite set of amino acid chemical structures from which evolution selected an alphabet of 20 building blocks for converting genetic instructions into living organisms. We found that the sub-set of amino acids used by biology exhibits some surprisingly simple, strikingly non-random properties if we use the Murchison meteorite as a guide to the amino acids that formed in a pre-biological Solar system.
In particular, a random collection of amino acids is highly unlikely to match the wide, even spread with which evolution’s “choice” represents molecular size, charge and hydrophobicity Philip and Freeland 2010. This feature of the amino acid alphabet appears to have been held more or less constant over the past 3 billion years – even during the earliest period, when life was evolving from an alphabet of ~8 amino acids into the standard 20 of modern biology. We have subsequently extended this analysis to show that the result holds for a wide variety of different scenarios for the molecular origins of life (manuscript in preparation). We have now started to find further experimental tests for this conclusion – such as a new study, led by UHNAI post-doctoral fellow J. Stephenson that asks whether biotechnological engineering efforts to extend, alter or simplify the biological amino acid alphabet match the conclusions we’ve drawn from physical chemistry.
More recently, we have started to apply this picture of the “functional logic” behind the amino acid alphabet to find bridges to the work of other NAI teams, particularly that led by Loren Williams at the Georgia Institute of Technology. Their work to reveal the evolutionary history of genetically encoded proteins reveals a natural, independent test for the conclusions that we are drawing from biophysical chemistry and evolutionary theory. In a related direction we have applied for NAI DDF funds to start a new collaboration with H. James Cleaves (Ga Tech.) to compute the ~27,000 amino acid structures that are of most relevance to life by virtue of similarity in their chemical structures.
During the year, Co-I Freeland has had the pleasure of presenting this work in person to: a group of 40 international graduate and post-doctoral researchers at the UH NAI Winter School, the NAI EC in-person meeting for PI’s, Johns Hopkins university, UMBC, and Georgia Institute of Technology. We have also presented this work via the web to Montana State U. NAI team, and as part of NAI’s first “workshop without walls.” Within the broader area of education and public outreach, Co-I Freeland has written a careful response to current claims by Intelligent Design that no natural process could have produced an origin for genetically coded information.
Noting that life uses a standard alphabet of just 20 amino acids to build proteins, despite the hundreds (if not thousands) that are possible, we investigated whether biological evolution appears to have favored some simple, non-random chemical properties.
We therefore defined an appropriate pool of plausible amino acids as a start point, and used computers to generate millions of alternative amino acid alphabets in order to measure their properties as a set of building blocks for proteins. We found that evolution’s choice outperforms random alternative selections for some simple, predictable criteria. This in turn gives us deeper insight into the relative roles of chance versus predictability in the emergence and evolution of life on our planet.
Publications
-
Philip, G. K., & Freeland, S. J. (2011). Did Evolution Select a Nonrandom “Alphabet” of Amino Acids?. Astrobiology, 11(3), 235–240. doi:10.1089/ast.2010.0567
- Freeland, S. (2011, In Press). The Origins of Genetic Information. Perspectives in Science and Christian Faith.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Stephen Freeland
Project Investigator
Gayle Philip
Collaborator
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.
Objective 4.3
Effects of extraterrestrial events upon the biosphere
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.2
Adaptation and evolution of life beyond Earth
Objective 7.1
Biosignatures to be sought in Solar System materials