2011 Annual Science Report
 Rensselaer Polytechnic Institute
Reporting | SEP 2010 – AUG 2011
Rensselaer Polytechnic Institute
Reporting | SEP 2010 – AUG 2011
Project 9: Microenvironmental Influences on Prebiotic Synthesis
Project Summary
Before biotic, i.e., “biologically-derived” pathways for the formation of essential biological molecules such as RNA, DNA and proteins could commence, prebiotic pathways were needed to form the molecules that were the basis for the earliest life. Much research has been done on possible non-biological routes to synthesis of RNA, thought by many to be the best candidate or model for the emergence of life. Our work focuses on possible physicochemical microenvironments on early earth that could have influenced and even directed or templated the formation of RNA or its predecessors.
Project Progress
Potential pitfalls in MALDI-MS analysis of abiotically produced RNA polymerization products
The MALDI-MS technique is much better characterized and optimized for analysis of proteins than for analysis of oligonucleotides. Interference from the MALDI matrix is not limited to background signal but may also promote formation of clusters of ribonucleotides. These clusters could be mistaken for actual polymerization products. Separate analysis by HPLC can confirm interpretation of MALDI-MS results but this is at the expense of the potentially superior detectability of MALDI-MS that could detect longer oligomers. We are performing systematic studies of MALDI-MS analysis of RNA polymerization products along with oligomer standards in order to improve the reliability of this approach for studying abiotic RNA synthesis.
Why G?
Regardless of whether RNA is the original “molecule of life” or evolved from less optimal precursors, it is extremely likely that it played a key role in the earliest stages of life on Earth. It is important to consider the ultimate selection of each of the components of RNA ribonucleotides (ribonucleobases C,A,U,G; ribose sugar, phosphate): why these particular nucleobases? Why ribose? Why phosphate? In particular, we are fascinated and puzzled by the inclusion of guanosine monophosphate (GMP) among the four ribonucleotides. This is because guanosine, GMP and other guanosine derivatives exhibit a unique property of reversible self-aggregation to form guanosine tetrads that can further associate through pi-pi stacking to form chiral, columnar aggregates and, at higher concentrations, lyotropic liquid crystalline phases. This alternate pathway for GMP competes with its incorporation into oligonucleotides, which is why it is difficult to synthesize highly G-rich RNA or DNA with good efficiency.
Given the competing pathway for GMP, we can ask if G is one of the four ribonucleotides in RNA in spite of, or because of, its unique properties. Our hypothesis is that the answer is “because of”. For one thing, G obviously has been quite successful in both RNA and DNA. But more intriguing is the possibility that G served a regulatory function that guided RNA sequences in a particular direction among the astronomically large combinatorial space of possibilities. This, combined with the ability of some G-rich sequences to form highly stable, intramolecular G-quadruplex structures that have been observed in aptamers, telomeres, and numerous G-rich sequences from genomic DNA, is the basis for our hypothesis.
As a first step toward testing our hypothesis, we are exploring the effect of GMP on the formation of RNA oligomerization by activated ribonucleotides (ImpX where X=A,G,C or U) in the presence of catalytic clays. The unactivated GMP should not itself be incorporated directly into the oligomers but its presence will increase the rate of the alternate pathway of G-tetrad formation. This could result in less G in the oligomers; however, it may instead serve as templating mechanism to guide the formation of oligomers with G-rich sequences.
There are many interesting factors to consider in this work, including the tendency of guanosine compounds to form highly viscous gels at high monomer concentrations, especially in the presence of cations, particularly K+ and Na+. One could imagine such gels reversibly and repeatedly forming and dissociating as a result of thermal or chemical cycles such as day-night cycles, tidal cycles, etc. The gels could form on bulk or particulate surfaces especially surfaces that offer favorable conditions of local pH and cations. For example, one might imagine a GMP-type film forming on catalytic particulates.
In our earliest experiments this year, we decided to investigate not only the effects of GMP alone but also in combination with the other ribonucleotides since they would be present in the prebiotic environment as well. Surprisingly, we discovered that in some combinations, the gel properties of GMP were significantly altered. We had already discovered the unique properties of gels formed by binary mixtures of guanosine with GMP but this is to our knowledge the first observation of gels formed by mixtures of GMP with other nucleotides. We are now conducting spectroscopic studies of the structure and thermal stability of these mixed gels. For example, the chart below summarizes the properties of several solutions of GMP with XMP=CMP.
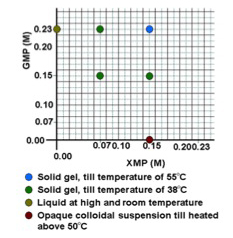
Fig.1. Chart summarizing the properties of several solutions of GMP with XMP=CMP.
We are also studying the ability of the gel solutions to suspend catalytic clays. We will use the results of these studies to guide the design of abiotic RNA synthesis experiments in solutions of the unactivated GMP alone and with XMP.
Effects of environmental conditions and components on abiotic RNA polymerization
Here we are interested in the effects of compounds that may have been present in the prebiotic environment during abiotic RNA synthesis. Our initial focus is on amino acids. We titrated montmorillonite clays with either alanine or tryptophan to determine any effects on RNA polymerization. We are in the process of analyzing the results using HPLC and MALDI-MS.
The work in this report forms the basis for the thesis research of NAI-supported graduate students Lauren Cassidy and Bradley Burcar.
Fig.2. The Prebiotic Chemistry Lab at RPI. Left to right: Bradley Burcar, Lauren Cassidy and Linda McGown.
Publications
- Burcar, B., Cassidy, L. & McGown, L.B. (2011). RNA Polymerization Using Catalytic Montmorillonite With Incorporated Amino Acids. AbGradCon 2011. Bozeman, MT.
- McGown, L.B. (2011). Sequence: A new dimension in rapid metagenomic profiling. Origins 2011 conference. Montpellier, France.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Prakash Joshi
Collaborator
Bradley Burcar
Graduate Student
Lauren Cassidy
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
