2011 Annual Science Report
 Montana State University
Reporting | SEP 2010 – AUG 2011
Montana State University
Reporting | SEP 2010 – AUG 2011
The Subglacial Biosphere – Insights Into Life-Sustaining Strategies in an Extraterrestrial Analog Environment
Project Summary
Sub-ice environments are prevalant on Earth today and are likely to have been more prevalent the Earth’s past during episodes of significant glacial advances (e.g., snow-ball Earth). Numerous metabolic strategies have been hypothesized to sustain life in sub-ice environments. Common among these hypotheses is that they are all independent of photosynthesis, and instead rely on chemical energy. Recently, we demonstrated the presence of an active assemblage of methanogens in the subglacial environment of an Alpine glacier (Boyd et al., 2010). The distribution of methanogens is narrowly constrained, due in part to the energetics of the reactions which support this functional class of organism (namely carbon dioxide reduction with hydrogen and acetate fermentation). Methanogens utilize a number of metalloenzymes that have active site clusters comprised of a unique array of metals. During the course of this study, we identified other features that were suggestive of other active and potentially relevant metabolic strategies in the subglacial environment, such as nitrogen cycling. The goals of this project are 1) identifying a suite of biomarkers indicative of biological CH4 production 2). quantifying the flux of CH4 from sub-ice systems and 3). developing an understanding how life thrives at the thermodynamic limits of life. This project represents a unique extension of the ABRC and bridges the research goals of several nodes, namely the JPL-Icy Worlds team and the ASU-Follow the Elements team.
Project Progress
Building upon our 2008/2009 work documenting the presence, diversity, and activity of methanogenic archaea in subglacial environments, we have now examined the abundance and physiology of methanogens that inhabit the subglacial sediments at Robertson Glacier, Alberta, Canada. Primers designed to specifically amplify the two predominant methanogen 16S rRNA gene phylotypes were used in qPCR reactions to quantify the abundance of each of these phylotypes in environmental samples collected in 2008, 2009, and 2010. In parallel, ex situ microcosms were initiated to identify the predominant mode of metabolism [e.g., acetoclastic, hydrogenotrophic, or methylotrophic] supporting the methanogen population in the subglacial environment, which when linked with our qPCR data directly allows us to link these uncharacterized populations with activity.
A collaboration between the ASU-Follow the Elements team (Everett Shock) was initiated in 2009 and has proven effective at leveraging and integrating the geochemical and thermodynamic expertise within the NAI with our expertise in microbial ecology. Since the initiation of our collaboration, we have generated comprehensive profiles of i). sediment porewater redox and trace element chemistry and ii). bulk meltwater and sediment C and N isotopic characterizations for samples collected in 2009, 2010, and 2011. Beginning in the fall of 2010, the ASU team also began collecting sediment porewater dissolved gas profiles in the field, and determined the presence of elevated concentrations of a number of relevant dissolved gases, including methane. These data are currently being integrated with biological data to predict the energetic feasibility of various metabolisms in the subglacial environment, including various forms of methanogenesis. These predictive data will in turn be used to evaluate the
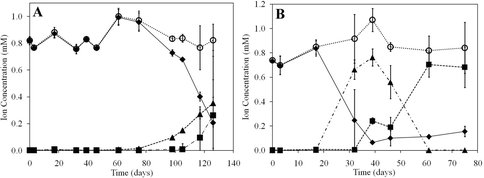
Concentrations of NH4+ (♦), NO2- (▲), NO3- (■), and total soluble N (○) as a function of time in microcosms incubated at 4°C (A) and 15°C (B) with 1 mM NH4+ added. Total soluble N is the sum of the concentrations of NH4+, NO2-, and NO3-. Values represent the difference in concentration between biological replicates and killed controls. The standard deviation of three replicates is indicated. These results for the first time demonstrate nitrification in the subglacial environment, a process that is likely to consume O2 and rendor the microhabitat anoxic. This is a requirement for active methanogenesis and points to a connection between the nitrogen and carbon cycle in subglacial enviorments. Figure adapted from Boyd et al., 2011. Appl. Environ. Microbiol. 77: 4778-4787.
Publications
-
Boyd, E. S., Lange, R. K., Mitchell, A. C., Havig, J. R., Hamilton, T. L., Lafreniere, M. J., … Skidmore, M. (2011). Diversity, Abundance, and Potential Activity of Nitrifying and Nitrate-Reducing Microbial Assemblages in a Subglacial Ecosystem. Applied and Environmental Microbiology, 77(14), 4778–4787. doi:10.1128/aem.00376-11
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
John Peters
Co-Investigator
Mark Skidmore
Co-Investigator
Trinity Hamilton
Doctoral Student
Matthew Urschel
Unspecified Role
-
RELATED OBJECTIVES:
Objective 2.1
Mars exploration.
Objective 2.2
Outer Solar System exploration
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 6.2
Adaptation and evolution of life beyond Earth
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems