2011 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2010 – AUG 2011
Massachusetts Institute of Technology
Reporting | SEP 2010 – AUG 2011
Origins of Multicellularity
Project Summary
By comparing animal genomes with genomes from their closest living relatives, the choanoflagellates, we can reconstruct the genome composition of the last common ancestor of animals.
Project Progress
To gain insight into the origin of animals, we aim to reconstruct the genome of the last common ancestor of animals and their sister group, the choanoflagellates. The ability of some choanoflagellates to form small multicelled colonies raises the possibility that their last common ancestor with animals was also transiently multicellular. To investigate the mechanisms and evolutionary history of transient multicellularity in choanoflagellates, the King lab has begun to develop the colony-forming choanoflagellate Salpingoeca rosetta into an experimentally tractable organism [1]. Furthermore, we have spent the last year (in collaboration with the Harvard/MIT Broad Institute) characterizing the genome of the colony forming choanoflagellate Salpingoeca rosetta, particularly focusing on what it reveals about the regulation of colony formation and the evolution of animal genomes.
By juxtaposing sponge and eumetazoan genomes with those from choanoflagellates, we have identified genes that were present in their common ancestor, as distinguished from genomic innovations that arose after their divergence. An earlier comparison using the genome of the single-celled choanoflagellate Monosiga brevicollis revealed that the seven major developmental signaling pathways of animals (NHR, Wnt, TGF-b, RTK, Notch, Hedgehog, and Jak-STAT) were present in the last common ancestor of animals while the RTK pathway, cadherins, C-type lectins, collagen, and components of the Hh and Jak/STAT signaling pathways evolved before the divergence of the choanoflagellate and animal lineages. With the incorporation of the S. rosetta genome into our analyses, we have been able to refine our reconstruction of the ancestral animal genome and have identified an additional set of transcription factors and signaling pathway components that evolved before the origin of animals. Two salient points may be drawn from these observations. First, the evolution of new signaling pathways along the metazoan lineage may have contributed to the emergence of bona fide animals. Second, the unicellular progenitor of animals contained abundant cell adhesion and signaling domains that first evolved in a pre-metazoan context. Our future research will aim to refine and expand our understanding of the content of ancestral genomes, while also revealing the role that protein domain shuffling and genome rearrangements might have contributed to the evolution of choanoflagellate and animal genomes.
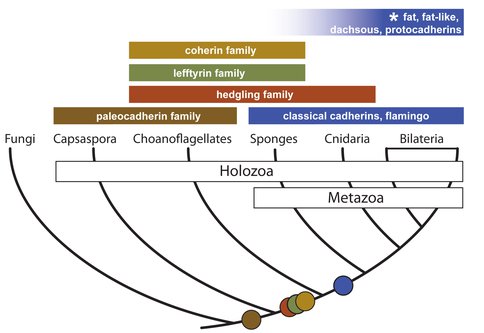
To better understand the evolution of multicellularity in animals, we have focused on cadherins, a family of transmembrane proteins required for animal cell adhesion, cell polarity, and multicellularity. Cadherins are also found in choanoflagellates, but the connection between cadherin evolution and animal origins is unknown. To reconstruct cadherin evolution, we analyzed the newly sequenced genomes of S. rosetta, the more distant animal relative Capsaspora owczarzaki, and the sponge Oscarella carmela. We have found that three new classes of cadherins — paleocadherins, lefftyrins, and coherins — evolved before the origin of animal and are absent from bilaterians. Paleocadherins are exclusive to C. owczarzaki and choanoflagellates, whereas lefftyrins and coherins are restricted to choanoflagellates and sponges. In contrast, classical cadherins, which interact with β -catenin to regulate cell adhesion in bilaterians, are found in sponges, but are absent from animal outgroups. Finally, we have found evidence for a conserved classical cadherin/ β-catenin complex in O. carmela, lending support to the hypothesis that classical cadherins were critical for animal origins.
References
1. Dayel, M.J., Alegado, R.A., Fairclough, S.R., Levin, T.C., Nichols, S.A., McDonald, K., and King, N. (2011). Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol 357, 73-82.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Scott Nichols
Postdoc
Daniel Richter
Graduate Student
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.