2011 Annual Science Report
 NASA Jet Propulsion Laboratory - Icy Worlds
Reporting | SEP 2010 – AUG 2011
NASA Jet Propulsion Laboratory - Icy Worlds
Reporting | SEP 2010 – AUG 2011
Detectability of Life
Project Summary
Detectability of Life investigates the detectability of chemical and biological signatures on the surface of icy worlds, with a focus on spectroscopic techniques, and on spectral bands that are not in some way connected to photosynthesis.Detectability of life investigation has three major objectives: Detection of Life in the Laboratory, Detection of Life in the Field, and Detection of Life from Orbit.
Project Progress
Overview
The primary component of Investigation 3 is the field campaign in Barrow, AK to characterize and quantify methane release from the Alaskan North Slope region and to understand the origin and fate of the methane. This work focuses on a handful of thermokarst lakes near Barrow and extends to the roughly 13,000 lakes throughout the region, an unknown fraction of which are release methane to the atmosphere.
Thermokarst lakes are topographic depressions created by thawing of ground ice, as a result of climate change over long periods of time or localized disturbances to the ground’s thermal regime (Davis, 2001). In the Northern Slope of Alaska thermokarst lakes are known to emit ebullient methane (CH4) some of which is of biogenic origin (Walter et al., 2007).
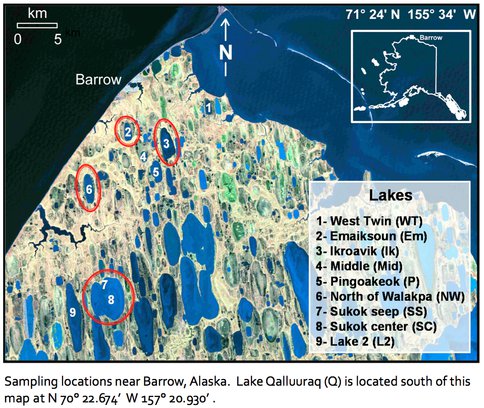
Figure 3-1. This figure shows the lakes at which our team has been working and from which samples have been collected for limnological and microbiological analyses.
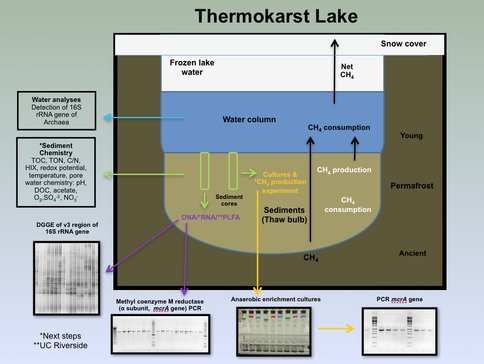
Figure 3-2. A cross-section diagram illustrating the various components of the lakes and the links to various science investigations is shown.
Key team members for this effort are Co-I’s Priscu (MSU), Murray (DRI/UNR), Hand (JPL), Love (UCR), and Walter-Anthony (UA Fairbanks). During the 2010/11 NAI effort our team continued lab analyses on samples from the April 2010 field campaign and conducted addition work and sampling during an October 2010 expedition.
Limnology and Methane Oxidation in Arctic Lakes
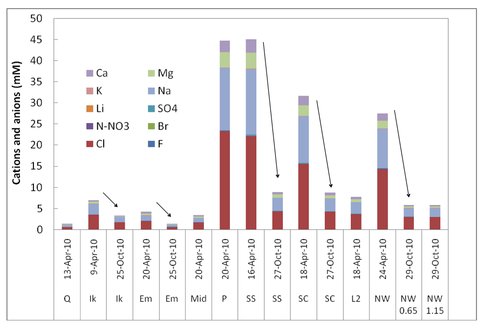
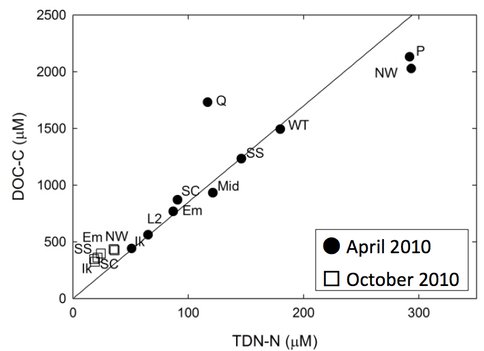
Microbes in the lakes of the North Slope near Barrow live in a wide range of ion and nutrient concentrations. Figure 3-3 shows a comparison of ions in different lakes and from Spring (max ice cover, minimum liquid water) to October (thin ice, freeze down just beginning). Carbon and nitrogen inventories as a function of Spring vs. Fall are shown in Figure 3-4 In this and subsequent figures the abbreviations given for the lakes are those given on Figure 3-1.
Figure 3-3. A comparison of ions in different lakes and from Spring (max ice cover, minimum liquid water) to October (thin ice, freeze down just beginning) is shown.
Figure 3-4. Carbon and nitrogen inventories as a function of Spring vs. Fall are shown.
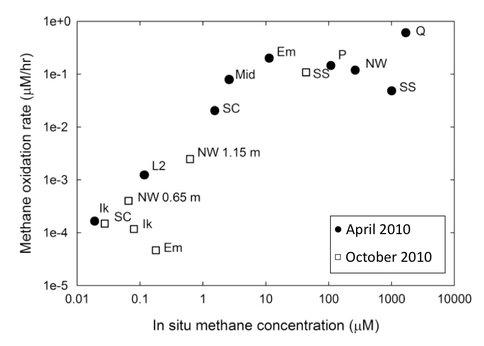
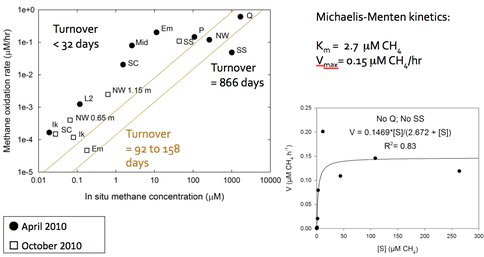
Methanotrophs in mixed communities are saturating in methane oxidation rates. Using this data (Figure 3-5) and rate kinetics calculations, combined with lab experiments on growth rates, Co-I Priscu and postdoc Adams have made an initial determination that the methanogenic biomass turnover time lies between 92-158 days, but may be as rapid as 32 days or less (Figure 3-6).
Figure 3-5. The relationship between measured methane concentrations and the rates at which the methanotrophs are utilizing that methane is shown. (Numbers next to some lakes indicate sampling depth.)
Figure 3-6. An initial determination of the methanogenic biomass turnover time is shown.
The methogen communities differ both temporally and spatially, with possible patterns emerging as a function of individual lake chemistry. The team is continuing analysis of DOM fluorescence data and pyrosequencing of bacterial communities is underway for select samples.
Archeal Communities and Methanogen Microbiology
The main goal of the Archeal work is to constrain the contribution of biological CH4 production to the CH4 budget of the thermokarst lakes. Our focus is on the community of Archaea that inhabit the sediments of these lakes, particularly those Archaea responsible for the production and consumption of CH4.
Two sampling campaigns took place in 2010, the first one in April, and the second one in October. During these campaigns 7 lakes (Ikroavik, Emaiksoun, Pingoakeok, Middle Lake, North of Walakpa, Sukok Seep and Center, and White Lake) around the Arctic Native village of Barrow, and Lake Qalluuraq, which is located further south, near the town of Atqasuk, were sampled to survey the water column and sediments for the presence of Archaea.
A combination of molecular microbiology tools are being used to study the community of Archaea in the water column and sediments of these lakes. Ten liters of water were collected from all the lakes in April, and from lakes Ikroavik, Sukok (Seep and Center sites), Emaiksoun and North of Walakpa in October. Water samples were filtered through 3 µm Versapor 3000 membrane filters (47 mm) and subsequently passed through 0.22 μm Sterivex filters. The samples collected this way were stored in a buffer at -80°C until community genomic DNA was extracted using a phenol:chloroform protocol.
Sediment cores were collected from lakes Ikroavik, Sukok (Seep and Center sites) and Qalluuraq in April, and lakes Ikroavik, Sukok (Seep) and North of Walakpa in October. In April, core lengths ranged between 17 and 74 cm, and sediment plugs were obtained from selected sections of the cores and preserved at -80°C. In October, core lengths ranged between 28 and 105 cm and the entire cores were kept frozen until segmentation and processing. One gram of sediment was later used for community genomic DNA extraction with the Power Lyzer Power Soil DNA Isolation Kit (MoBio).
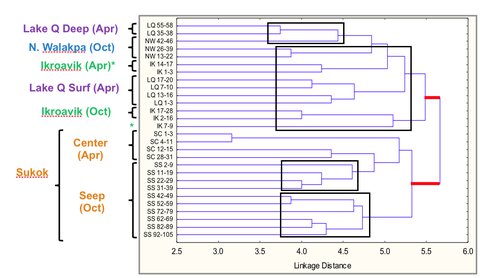
Archaea were detected by polymerase chain reaction (PCR) in all lake sediments at all depths sampled, using a few different combinations of primers for the 16S rRNA gene. However, Archaea have not been detected in the water column using the same PCR conditions for the sediment samples or a nested PCR approach. Profiling of the variable region 3 (v3) of the 16S rRNA of Archaea was carried out by denaturing gradient gel electrophoresis (DGGE). The v3 profiles indicated surprisingly high levels of diversity, with 18 to 43 bands per depth sampled. A clustering analysis (Figure 3-7) showed that at a linkage distance of 5.0 there were 8 clusters, indicative of partitioning of archaeal communities between lakes. Additionally, one of v3 rRNA gene bands was common to all lakes, and most phylotypes were grouped by depth (1-40 cm or 41-105 cm) within a lake.
Figure 3-7. Clustering analysis of Archea phylotypes obtained by DGCE profiling of the v3 region of the 16S rRNA gene is shown. Note that Sukok formed a separate cluster, and in general phylotypes grouped by depth: surface samples (1-40 cm) vs. deeper samples (41-105 cm).
To specifically look for methanogens and anaerobic methanotrophs (ANME) in the sediments, DNA was surveyed by PCR to detect the methyl coenzyme M reductase (mcrA) gene, which is specific to the pathways of methanogenesis and anaerobic methane oxidation (AMO). The expected 464-491 bp amplification product predicted for the mcrA gene was detected in all the sediment samples. Additionally, an array of methanogen enrichment cultures was set up in collaboration with Dr. Jeremy Dodsworth in Dr. Brian Hedlund’s lab at the University of Nevada Las Vegas, and the different media were inoculated with sediment slurries prepared anaerobically in autoclaved lake water. Enrichment cultures that were started after the first and second sampling campaigns contain substrates that are used in the main pathways for methanogenesis (acetate fermentation, carbon dioxide (CO2) reduction, and one-carbon dismutation). Forty-five cultures were started from the same depths sampled for DNA profiling in April and they have been incubated at 2°C, while in October separate cores were obtained for this purpose. The October cores were kept at 2-4°C until sectioning in ~ 30 cm fragments and then used as inocula for 54 cultures. These cultures have been incubated at 2 and 10°C.
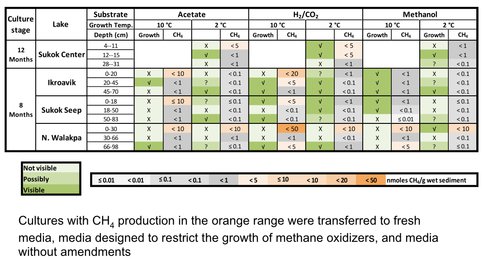
After 8 to 12 months of incubation, most cultures have shown signatures of growth (Figure 3-8). Many of these cultures have produced CH4 from CO2 reduction with hydrogen (H2), but a few others utilized methanol or acetate as methanogenic substrates (Figure 3-8).
Figure 3-8. Visible growth and methane production of enrichment cultures from Artic Lakes is demonstrated.
The cultures that produced CH4 were surveyed for the presence of the mcrA gene with positive results. The same cultures were also transferred to fresh medium and to new kinds of media designed to restrict the growth of Bacteria and methanotrophs. New control media was also designed to test for the use of methanogenic substrates and other possible sources of carbon. Moreover, a set of cultures is being incubated at 22°C to test for the presence of psychrotrophs, as operationally, psychrophiles do not grow over 20°C. The new culture set encompasses 85 cultures.
From the experiments conducted to date we conclude that there is a great diversity of Archaea inhabiting these thermokarst lakes, which include, in part, a methanogenic community. These findings suggest that the archaeal community in lake sediments of the Northern Slope of Alaska may be more relevant to estimates of methane release from the Arctic than previously thought. Our work will set the grounds to further the understanding of the effect of temperature increases on microbial activities that directly affect the greenhouse gas inventory, and it will expand the census of psychrophiles that thrive in permafrost environments and that could be used for testing under simulated Europa-like conditions. In the near future, we will collect fresh samples to perform activity analyses, including quantification of the rate of methane production in sediment samples from Lakes Sukok and North of Walakpa; RNA extraction for gene expression studies of the 16S rRNA and the mcrA genes; and pyrosequencing of the v3 region of the 16S rRNA gene from a few samples.
Methane Mapping and Detection
As part of our work to identify and map lakes that may be releasing large quantities of methane, we have been using data from the Moderate Resolution Imaging Spectroradiometers (MODIS) onboard the Terra and Aqua satellites. These satellites get near daily coverage of the Alaskan North Slope and one of the data products is a snow and ice coverage map for the region. Much of the region is often cloudy and the data is therefore not useful, but Co-I Hand gave an undergraduate intern from Brown the task of sifting through the data from the past ~10 years to look for freezing anomalies in the lake ice and snow cover that might be usable as a proxy for areas of intense and sustained methane bubbling.
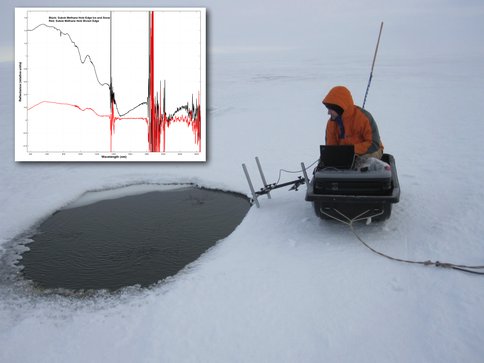
The basic concept came from the team’s experience at Sukok lake in October of 2010. We were conducting attempts to characterize the methane in the infrared via the 1.66 and 2.13 micron bands, but wind and weather hampered direct detection of the methane at the source. As Figures 3-9 and 3-10 show however, the methane seeps maintained open water regions through kinetic inhibition of freezing and therefore such open water or non-snow covered lake ice regions – were they large enough – could be observable from orbit with the MODIS data during the month of October when freeze over occurs.
Figure 3-9. Open water or non-snow covered regions of Sukok lake is shown during our in October of 2010 field expedition.
Figure 3-10. An investigator is having a closer look at an open water region of Sukok Lake.
The MODIS pixels, once corrected for latitude, span roughly 600-800 m/pixel and thus the resolution is very low, making the prospect of success more difficult. Nevertheless, Andrew Park, the talented intern, scoured the October data from 2002-2010 and generated maps and GPS points for future investigation. Figures 3-11 show an example of these maps. The light grey dotted points surrounded by white are lake ice and possible open water. In some cases the identified points correspond to larger lakes and thus the anomaly could be explained by thermal mass, but in other cases the points correspond to small lakes and methane seeping open water is a viable hypothesis. Future work will aim to ground truth these data points, helping to extend the methane mapping from the locale to the regional scale.
Figure 3-11. Maps and GPS points have been generated using the 2002 MODIS data to locate possible open water or non-snow covered lake ice regions for future investigations.
Remote Sensing of the Sierra Snowpack and Algae Blooms
As part of our continuing effort to map and understand the microbial biomass in the Sierra snowpack, Co-I’s Painter and Hand gathered data from several piggy-back flybys on the Airborne Visible and Infrared Imaging Spectrometer (AVIRIS). The primary mission was for a different spectroscopy investigation, but working with Collaborator Mike Eastwood our team was able to retrieve spectacularly clear data from AVIRIS showing the Sierra snowpack in July of 2011 (Figure 3-12). Investigators Hand and Painter are continuing to work on the data reduction and analyses.
Figure 3-12. The groundtracks and two examples of the quicklook data products from AVIRIS are shown.
References
Davis, N. 2001. Permafrost. A guide to frozen ground in transition. University of Alaska Press, Fairbanks, Alaska. pp. 351.
Walter, K. M., L. C. Smith, and F. S. Chapin III. 2007. Methane bubbling from northern lakes: present and future contributions to the global methane budget. Philosophical Transactions of the Royal Society A, 365(1856): 1657-1676
Walter, K. M., J. P. Chanton, F. S. Chapin, III, E. A. G. Schuur, and S. A. Zimov. 2008. Methane production and bubble emissions from arctic lakes: Isotopic implications for source pathways and ages. Journal of Geophysical Research, 113, G00A08,doi:10.1029/2007JG000569.
Publications
-
Dalton, J. B., Cruikshank, D. P., Stephan, K., McCord, T. B., Coustenis, A., Carlson, R. W., & Coradini, A. (2010). Chemical Composition of Icy Satellite Surfaces. Space Sci Rev, 153(1-4), 113–154. doi:10.1007/s11214-010-9665-8
-
Hand, K. P., & Carlson, R. W. (2011). H2O2 production by high-energy electrons on icy satellites as a function of surface temperature and electron flux. Icarus, 215(1), 226–233. doi:10.1016/j.icarus.2011.06.031
-
Hand, K. P., & Carlson, R. W. (2012). Laboratory spectroscopic analyses of electron irradiated alkanes and alkenes in solar system ices. Journal of Geophysical Research: Planets, 117(E3), n/a–n/a. doi:10.1029/2011je003888
-
Hand, K. P., Khurana, K. K., & Chyba, C. F. (2011). Joule heating of the south polar terrain on Enceladus. Journal of Geophysical Research, 116(E4), None. doi:10.1029/2010je003776
-
Ivanov, M. A., Prockter, L. M., & Dalton, B. (2011). Landforms of Europa and selection of landing sites. Advances in Space Research, 48(4), 661–677. doi:10.1016/j.asr.2011.05.016
-
Korablev, O., Gerasimov, M., Brad Dalton, J., Hand, K., Lebreton, J-P., & Webster, C. (2011). Methods and measurements to assess physical and geochemical conditions at the surface of Europa. Advances in Space Research, 48(4), 702–717. doi:10.1016/j.asr.2010.12.010
-
Shirley, J. H., Dalton, J. B., Prockter, L. M., & Kamp, L. W. (2010). Europa’s ridged plains and smooth low albedo plains: Distinctive compositions and compositional gradients at the leading side–trailing side boundary. Icarus, 210(1), 358–384. doi:10.1016/j.icarus.2010.06.018
- Adams, H.E. & Priscu, J.C. (2011). Bacterial activity in methane-rich, icy habitats. American Society for Limnology and Oceanography, Aquatic Sciences Meeting. San Juan, Puerto Rico.
- Adams, H.E. & Priscu, J.C. (2011, In Preparation). Methane oxidation in icy habitats.
- Adams, H.E., Hand, K.P., Murray, A.E., Lyons, B. & Priscu, J.C. (In Preparation). Distribution of bacterial communities, activity, and carbon sources in Alaskan coastal plain lakes.
- Dalton, J.B., Shirley, J.H. & Prockter, L.M. (2010). Surface geology of Europa: A window to subsurface composition and habitability. Geophysical Research Abstracts, 12.
- Hand, K.P. & Painter, T.H. (2010). Fall AGU Mtg.
- Hand, K.P. (2011). On the Coming Age of Ocean Exploration. In: Brockman, M. (Eds.). Future Science.
- Hand, K.P. Outer planet satellites as potential crucibles for life: extraterrestrial and terrestrial. UN meeting.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Heather Adams
Collaborator
Dan Berisford
Research Staff
Paula Matheus-Carnevali
Graduate Student
Megan Rohrssen
Graduate Student
Pamela Santibanez
Graduate Student
-
RELATED OBJECTIVES:
Objective 1.2
Indirect and direct astronomical observations of extrasolar habitable planets.
Objective 2.1
Mars exploration.
Objective 2.2
Outer Solar System exploration
Objective 4.1
Earth's early biosphere.
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 6.2
Adaptation and evolution of life beyond Earth
Objective 7.1
Biosignatures to be sought in Solar System materials
Objective 7.2
Biosignatures to be sought in nearby planetary systems