2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
High Level Theory - the Role of Mg2+ in Ribosome Assembly
Project Summary
Magnesium plays a special role in RNA function and folding. Although water is magnesium’s most common first-shell ligand, magnesium has significant affinity for the oxyanions of RNA phosphates. Here we provide a quantum mechanical (QM) description of first shell RNA-magnesium and DNA-magnesium interactions, demonstrating unique features that appear to be required for folding of large RNAs. Our work focuses on multidentate chelation of magnesium by RNA and DNA, where multiple phosphate oxyanions enter the first coordination shell of magnesium. The results suggest that magnesium, compared to calcium and sodium, has enhanced ability to form bidentate chelation complexes with RNA. Sodium complexes, in particular, are unstable and spontaneously open. A magnesium cation is closer to the oxyanions of RNA than the other cations, and is stabilized not only by electrostatic interaction with the oxyanions but also by charge transfer and polarization interactions. Those interactions are quite substantial at close distances. The quantum effects are less pronounced for calcium due to its larger size, and for sodium due to its smaller charge. Additionally, we find that magnesium complexes with RNA are more stable than those with DNA. The nature of the additional stability is twofold: it is due to a slightly greater energetic penalty of ring closure to form chelation complexes for DNA, and elevated electrostatic interactions between the RNA and cations. In sum it can be seen that even at high concentration, sodium and calcium cannot replicate the structures or energetics of RNA-magnesium complexes.
Project Progress
Magnesium plays a special role in folding of large globular RNAs (1, 2). These RNAs are closely associated with magnesium ions, many of which can be visualized by X-ray diffraction. For example, 118 magnesium ions associate with the 23S rRNA in the large ribosomal subunit of Haloarcula marismortui (PDB entry 1JJ2, described in 3, 4). Although magnesium’s most common first-shell ligand is water, the oxyanions of RNA phosphates (i.e., the non-bridging phosphate oxygens) have significant affinity for magnesium. Ninety-eight of the magnesium ions associated with the LSU of H. marismortui contain phosphate oxyanions within their first coordination shells (characterized by Mg2+-OP distances < 2.4 Å). Other RNAs show the same trend. Seventy-one magnesium ions associate with the P4-P6 domain of the tetrahymena Group 1 intron (PDB 1HR2, described in 5, 6). Twenty-six of these contain phosphate oxyanions within their first coordination shell.
Our work here focuses on multidentate chelation of magnesium by RNA, where multiple phosphate oxyanions enter the first coordination shell of a magnesium ion. These complexes are rigid with well-defined geometry because the first coordination shell of magnesium is a well-packed octahedron (7, 8). Tight ligand packing and crowding is a hallmark of magnesium complexes, leading to highly restrained geometry, and strong ligand-ligand interactions. Multidentate chelation of magnesium by RNA is also associated with non-canonical conformation states (1, 4, 9).
The most frequent multidentate chelation mode of magnesium by large RNAs is the coordination of a common cation by oxyanions of two adjacent nucleotides (1, 9) (Figure 1A, also see the supplementary information). One observes these “clamps” of adjacent phosphates on a magnesium ion twenty-five times in the H. marisortui large ribosomal subunit (3), twice in the P4-P6 domain of the tetrahymena Group 1 intron (5, 6), once in a self-splicing group II intron from Oceanobacillus iheyensis (PDB entry 3IGI, described in 10), and once in the L1 ligase (PDB entry 2OIU, described in 11). The folding and function of each of these RNAs is magnesium-dependent. The 10-membered ring systems (Mg2+-O-P-P-O5’-C5’-C4’-C3’-O3’-P-O-P-Mg2+, Figure 1A) that characterize these clamps appear to be elemental units of RNA folding and assembly. Tri- and tetradentate RNA-magnesium complexes nearly always contain at least one of these 10-membered ring systems (1). One magnesium ion is clamped twice, by both the mRNA and the rRNA, in the assembled Thermus thermophilus ribosome (PDB entry 2J01, described in 12).
The forces and energetics of cation association with nucleic acids and other ligands can be characterized by application of high level theory (13-16) using density functional theory methods (17-19). Here we provide a quantum mechanical (QM) description of first shell RNA-magnesium interactions, demonstrating unique features that appear to explain folding of large RNAs. The results here show that magnesium can induce specific conformational and electronic states of RNA that are inaccessible with other biological cations. The stability of complexes is dependent on cation type, position, and coordination, and has significant polarization, charge-transfer and exchange components. One must treat these systems quantum mechanically because continuum theories such as NLPB or GBSA are not applicable when cations are effectively part of the macromolecule (20). In cases where immobilized cations are responsible for specific structural integrity, they must be considered explicitly, not in a continuum framework.
Here we dissect the conformations and energetics of RNA and DNA clamps of magnesium, calcium, or sodium, in an aqueous environment. A magnesium-RNA clamp (Figure 1A) extracted from within the H. marismortui large ribosomal subunit (PDB entry 1JJ2) was used to build a template clamp containing phosphates attached to both the O3’ and O5’ atoms of a ribose. The 5’ and 3’ phosphates were capped with methyl groups in lieu of the remainder of the RNA polymer.
Magnesium, from the time of life’s origins, has been closely associated with some of the central players in biological systems – phosphates and phosphate esters (21). Here we show that magnesium shares a special geometric and energetic relationship with the phosphates of RNA. Adjacent RNA phosphates form clamps selectively with magnesium ions (Figure 1). The ionic radius of Mg2+ is small (0.65 Å), the charge density is high, the coodination geometry is octahedral (the AOCN, Average Observed Coordination Number, is 5.98), the preferred ligands are charged or neutral oxygens, and the hydration enthalpy is large (-458 kcal/mol) (7, 8, 22, 23). In comparison with group I ions, calcium, or polyamines, magnesium has a much greater affinity for phosphate oxygens, and binds to them with well-defined geometry. Unlike other cations, magnesium can bring phosphate oxygens in its first shell into close proximity.
Here we demonstrate that neither sodium nor calcium can replicate the structures or energetics of RNA-magnesium clamps. RNA forms more stable clamps with magnesium than with calcium or sodium. Clamps with sodium in particular are unstable, and spontaneously open. Magnesium is closer than the other cations to oxyanion ligands, and so magnesium clamps are stabilized not only by electrostatic interactions but also by charge transfer and polarization. Those interactions are quite substantial at short range. These effects are less pronounced for calcium due to its larger size and for sodium due to its smaller charge. The clamps are specific to RNA in that ribose clamps are more stable than deoxyribose clamps. The nature of the extra stability of RNA clamps is twofold: a) a slightly attenuated energetic penalty of the ring closure, and b) elevated electrostatic interactions between the RNA and cations. Thus, we can now explain, at least in part, the origin of the special role of magnesium in folding of large RNAs.
References
1. Hsiao C, Tannenbaum M, VanDeusen H, Hershkovitz E, Perng G, Tannenbaum A, Williams LD (2008) Complexes of Nucleic Acids with Group I and II Cations. Nucleic Acid Metal Ion Interactions, ed Hud N (The Royal Society of Chemistry, London), pp 1-35.
2. Draper DE, Grilley D, Soto AM (2005) Ions and RNA Folding. Annu Rev Biophys Biomol Struct 34:221-243.
3. Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science 289:905-920.
4. Klein DJ, Moore PB, Steitz TA (2004) The Contribution of Metal Ions to the Structural Stability of the Large Ribosomal Subunit. RNA 10:1366-1379.
5. Juneau K, Podell E, Harrington DJ, Cech TR (2001) Structural Basis of the Enhanced Stability of a Mutant Ribozyme Domain and a Detailed View of RNA—Solvent Interactions. Structure 9:221-231.
6. Cate JH, Hanna RL, Doudna JA (1997) A Magnesium Ion Core at the Heart of a Ribozyme Domain. Nat Struct Biol 4:553-558.
7. Brown ID (1992) Chemical and Steric Constraints in Inorganic Solids. Acta Crystallogr Sect B 48:553-572.
8. Bock CW, Katz AK, Markham GD, Glusker JP (1999) Manganese as a Replacement for Magnesium and Zinc: Functional Comparison of the Divalent Ions. J Am Chem Soc 121:7360-7372.
9. Hsiao C, Williams LD (2009) A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res 37:3134-3142.
10. Toor N, Keating KS, Taylor SD, Pyle AM (2008) Crystal Structure of a Self-Spliced Group II Intron. Science 320:77-82.
11. Robertson MP, Scott WG (2007) The Structural Basis of Ribozyme-Catalyzed RNA Assembly. Science 315:1549-1553.
12. Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S Ribosome Complexed with mRNA and tRNA. Science 313:1935-1942.
13. Rulisek L, Sponer J (2003) Outer-Shell and Inner-Shell Coordination of Phosphate Group to Hydrated Metal Ions (Mg2+, Cu2+, Zn2+, Cd2+) in the Presence and Absence of Nucleobase. The Role of Nonelectrostatic Effects. J Phys Chem B 107:1913-1923.
14. Gresh N, Sponer JE, Spackova N, Leszczynski J, Sponer J (2003) Theoretical Study of Binding of Hydrated Zn(II) and Mg(II) Cations to 5’-Guanosine Monophosphate. Toward Polarizable Molecular Mechanics for DNA and RNA. J Phys Chem B 107:8669-8681.
15. Munoz J, Sponer J, Hobza P, Orozco M, Luque FJ (2001) Interactions of Hydrated Mg2+ Cation with Bases, Base Pairs, and Nucleotides. Electron Topology, Natural Bond Orbital, Electrostatic, and Vibrational Study. J Phys Chem B 105:6051-6060.
16. Trachtman M, Markham GD, Glusker JP, George P, Bock CW (1998) Interactions of Metal Ions with Water: Ab Initio Molecular Orbital Studies of Structure, Bonding Enthalpies, Vibrational Frequencies and Charge Distributions. 1. Monohydrates. Inorg Chem 37:4421-4431.
17. Murashov VV, Leszczynski J (1999) Theoretical Study of Complexation of Phosphodiester Linkage with Alkali and Alkaline-Earth Cations. J Phys Chem B 103:8391-8397.
18. Petrov AS, Lamm G, Pack GR (2005) Calculation of the Binding Free Energy for Magnesium-RNA Interactions. Biopolymers 77:137-154.
19. Petrov AS, Pack GR, Lamm G (2004) Calculations of Magnesium-Nucleic Acid Site Binding in Solution. J Phys Chem B 108:6072-6081.
20. Schurr JM (2008) Polyanion Models of Nucleic Acid-Metal Ion Interactions. Nucleic Acid Metal Ion Interactions:201-244.
21. Westheimer FH (1987) Why Nature Chose Phosphates. Science 235:1173-1178.
22. Brown ID (1988) What Factors Determine Cation Coordination Numbers. Acta Crystallogr Sect B 44:545-553.
23. Rashin AA, Honig B (1985) Reevaluation of the Born Model of Ion Hydration. J Phys Chem 89:5588-5593.
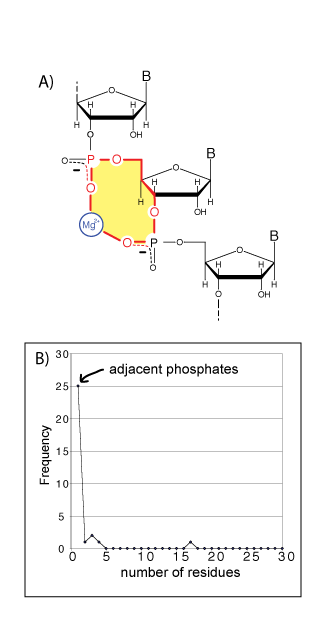
A) A schematic diagram of a bidentate RNA clamp of magnesium, formed when adjacent phosphate groups enter the first coordination shell of a common magnesium ion. A 10-membered ring (shaded yellow) characterizes these bidentate RNA clamps. B) Adjacent phosphate groups enter the first coordination sphere of a common phosphate more commonly than non-adjacent phosphates. A bidentate RNA clamp of magnesium corresponds to one residue in the frequency graph below (frequency from the H. marismortui 23S rRNA, PDB entry 1JJ2). Extending the horizontal axis (not shown) reveals local phosphates enter the first shell of a common magnesium ion preferentially over remote phosphates.
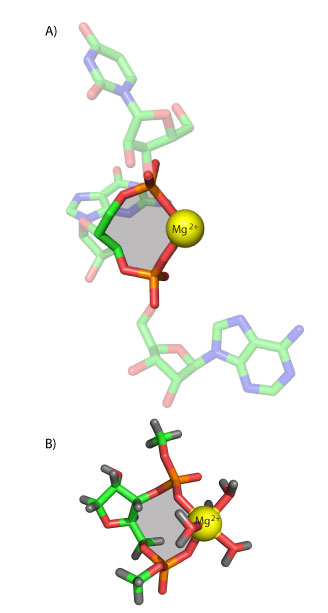
A) A bidentate RNA clamp of magnesium observed in the H. marismortui ribosomal LSU crystal structure (Mg 8003 from PDB entry 1JJ2). B) The RNA2—Mg2+.•(H2O)4 complex used here for QM calculations. The base has been replaced by a hydrogen atom and the chain is terminated with methyl groups. Ten-membered rings are shaded yellow.
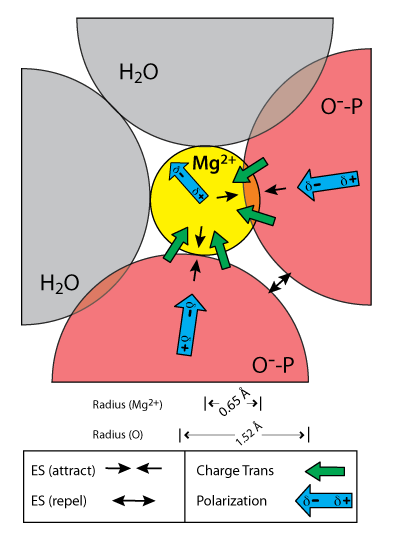
Interactions of a hexacoordinated magnesium ion with two anionic oxygen atoms and four water molecules (only the two in-plane water molecules are shown). Illustrated distances and angles are based on the QM calculations. Arrows represent electrostatic, polarization, and charge transfer components of the interaction energy as obtained by NEDA. Only the major components of the interaction energy are shown. The exchange term, which is favorable but significantly weaker than the corresponding charge transfer and polarization terms, is omitted from the schematic diagram for clarity.
Publications
-
Petrov, A. S., Bowman, J. C., Harvey, S. C., & Williams, L. D. (2010). Bidentate RNA-magnesium clamps: On the origin of the special role of magnesium in RNA folding. RNA, 17(2), 291–297. doi:10.1261/rna.2390311
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Anton Petrov
Postdoc
Jessica Bowman
Research Staff
Chad Bernier
Graduate Student
Denise Enekwa
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 4.2
Production of complex life.


