2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
An Atomic Level Description of the Specific Interactions Between Nascent Peptide and Ribosome Exit Tunnel
Project Summary
The ribosome exit tunnel is an ancient path that must be traveled by all peptides/proteins synthesized by the ribosome. We have synthesized peptolides and demonstrated their potential as probes to decipher the interaction between the nascent peptide and the exit tunnel. This study is yielding vital information about attributes that confer nucleic acids with selective advantage as building blocks for exit tunnel construction.
Project Progress
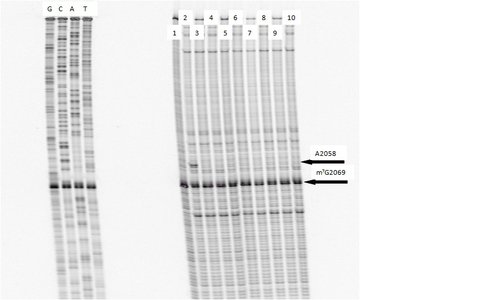
The ribosome, through highly coordinated and concerted movement, is responsible for the synthesis of all protein found within a cell. In addition, it is so highly conserved that is can serve as a molecular time capsule, varying subtlety within the tree of life. While new technology has led to an explosion of knowledge concerning the components of the ribosome, many questions remain. One of these questions involves the nature of the exit tunnel and its level of interaction with a growing peptide before breeching the extraribosomal interface. Through the use of peptolides (ketolide-peptide hybrids described previously), we report a level of interaction between varied peptide sequences and key residues within the exit tunnel. This is being accomplished by cell-free translation inhibition assays followed by footprinting experiments of the ribosomal RNA (rRNA) through dimethyl sulfate (DMS) and 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) modification in the presence of the peptolides. Translation inhibition experiments, which showed that antibiotic activity of the peptolides remained intact (Table 1), have been finalized. With the knowledge that the peptolides retain functionality, a series of primers and conditions have been optimized for footprinting experiments. Optimization was done for dideoxy termination (Sanger) sequencing, reverse transcription, as well as gel electrophoresis conditions (1). The first interaction to target through footprinting was A2058 (E. coli numbering). This key residue has been shown through numerous crystallographic models to serve as the binding site for macrolides and ketolides (2-6). As shown in Figure 1, A2058 is strongly protected from modification by DMS by our peptolides. Preliminary results are suggesting strong levels of interactions at multiple residues on the rRNA that can be seen in Figures 2 and 3. Experimentation is ongoing to collect data for DMS and CMCT footprinting on all four primers, while looking for additional regions to explore by yet to be determined primers.
References
1. Merryman, C & Noller, H. RNA-Protein Interactions: a Practical Approach. Oxford University Press, New York, 1998.
2. Petrepoulos, A. D., Kouvela, A.C., Starosta, A.L., Wilson, D.N., Dinos, G.P., Kalpaxis, D.L. J Mol Biol. 2009. 385, 1179-1192.
3. David-Eden, H., Mankin, A.S., Mandel-Gutfreund, Y. NAR. 2010. 38(18), 5982-5994.
4. Kannan, K., Mankin, A.S. N.Y. Acad. Sci. 2011. 1241, 33-47.
5. Ramu, H., Vazquez-Laslop, N., Klepacki, D., Dai, Q., Piccirilli, J., Micura, R., Mankin, A.S. Mol. Cell. 2011. 41, 321-330.
6. Mankin, A. TRENDS in Biochemical Studies. 2006, 31(1), 11-13.
Footprinting of 23S rRNA with primer 2180. Dideoxy sequencing lanes G,C,A and T followed by lanes: 1) unmodified rRNA, 2) DMS modified rRNA, Lanes 3-10) rRNA DMS modified in the presence of Clarithromycin, ketolide-azide, 3C NLS, 4C NLS, 3C rNLS, 4C rNLS, 3C PLI, and 4C PLI respectively.
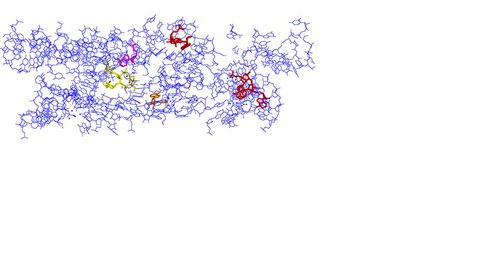
View of the exit tunnel from PDB: 3OFQ (20 Å diameter) with peptidyl transferase center on the left. Color coding as follows: Yellow = Erythromycin; Purple = A2058; Red = A1264, A1275, A1336; Orange = A751.
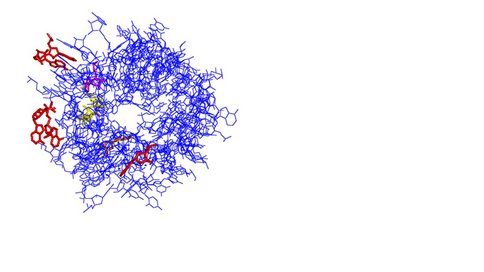
View through the exit tunnel from PDB: 3OFQ (20 Å diameter) looking back to the PTC. Color coding as follows: Yellow = Erythromycin; Purple = A2058; Red = A1264, A1275, A1336; Orange = A751.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Josh Canzoneri
Graduate Student
Arren Washington
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules