2011 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
Georgia Institute of Technology
Reporting | SEP 2010 – AUG 2011
A Phylogenomic Approach to the Ur-Ribosome
Project Summary
A macromolecular complex emulating ancient ribosomal function would sharpen understanding of the peptide bond’s emergence in a biological context. Reconstructing evolutionary adaptive paths (1) offers a straightforward approach to building an ancient ribosome, in principle: the sequences of the ribosomal proteins and ribosomal RNA are aligned, and subjected to likelihood-based phylogenetic reconstruction, followed by over-expression and purification of the ancestral components. In practice, sequence retrieval is complicated by inconsistent annotation, gene absence, and database redundancy. One accomplishment of the past year was development and implementation of a novel algorithm for large-scale protein sequence retrieval and family discernment.
Project Progress
Initial sequence retrievals found inconsistent sequence annotation between Archaeal, Bacterial, and Eukaryal rProtein families. For example, Bacterial L33 is orthologous to the L44/L36 family in Eukaryotes and Archaea. The family known as L29P in Bacteria and Archaea is orthologous to L35E in Eukaryota. Prokaryotic L14 shares a common ancestor with Eukaryotic L23E. Family L15 in Bacteria is orthologous to L15P in Archaea and L15/L27 in Eukaryota. Eukaryotic sequences are necessary to root Bacteria when gene loss has occurred in Archaea, however the organellar rProteins, and additional cytosolic paralogs, further complicate retrieval. We overcame these difficulties by developing a dynamic algorithm for Selectable Taxon Ortholog Retrieval Modestly (STORM). We use STORM to retrieve sequences, cull database redundancy, and delineate protein families within the context of evolutionarily related paralogs.
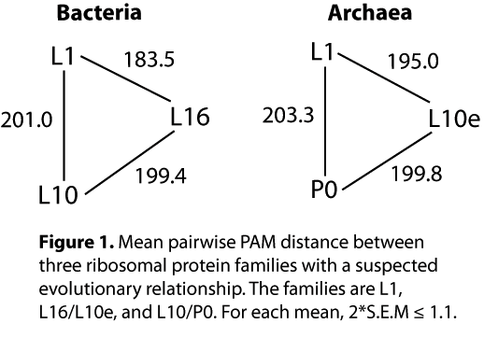
Although ribosomal paralogs are rare, there are reports of recent duplications in Bacteria (1) and numerous duplications in Eukaryota (2). Several authors note the L18e/L15P paralogy in Archaea, and Wang et al. (3) suggest paralogy of Archaeal L1 and L10e. Our preliminary analysis supports the latter, as well as the relatedness of L1, L10e, and P0. These Archaeal families have Bacterial orthologs with an apparently congruent evolutionary relationship (Fig. 1). The universal families L1, L16/L10e, and L10/P0 originated prior to the radiation of Life’s three domains. During the time of LUCA, ancestral L10 coexisted with a single protein that was the progenitor to L1 and L16/L10e. This phylogenetic reconstruction supports and complements reconstruction from assembly-dependency data (4), wherein L10 is prior to L16 but the priority of L1 is unresolved because it binds to the intermediate ribosomal particle independently of other rProteins. Assuming that a) the present families share a common ancestor, and b) their evolutionary rate is clock-like, we tentatively conclude that L1 emerged concurrently with L16/L10e, after emergence of the L10/P0 ancestor. Ongoing and future work explores assumptions (a) and (b), employing more detailed phylogenetic analysis (5), structural modeling (6) and in vivo resurrection (7).
References
1. Makarova KS, Ponomarev VA, Koonin EV (2001) Two C or Not Two C: Recurrent Disruption of Zn-Ribbons, Gene Duplication, Lineage-Specific Gene Loss, and Horizontal Gene Transfer in Evolution of Bacterial Ribosomal Proteins. Genome Biol 2:RESEARCH 0033.
2. Lecompte O, Ripp R, Thierry JC, Moras D, Poch O (2002) Comparative Analysis of Ribosomal Proteins in Complete Genomes: An Example of Reductive Evolution at the Domain Scale. Nucleic Acids Res 30:5382-5390.
3. Wang J, Dasgupta I, Fox GE (2009) Many Nonuniversal Archaeal Ribosomal Proteins Are Found in Conserved Gene Clusters. Archaea (Vancouver, BC) 2:241-251.
4. Fox GE, Ashinikumar KN (2004) The Evolutionary History of the Translation Machinery. The Genetic Code and the Origin of Life, ed de Pouplana LR (Kluwer Academic / Plenum Publishers, New York ), pp 92-105.
5. Zhao ZM, Reynolds AB, Gaucher EA (2011) The Evolutionary History of the Catenin Gene Family During Metazoan Evolution. BMC Evol Biol 11:198.
6. Roy A, Kucukural A, Zhang Y (2010) I-Tasser: A Unified Platform for Automated Protein Structure and Function Prediction. Nature protocols 5:725-738.
7. Gaucher EA, Govindarajan S, Ganesh OK (2008) Palaeotemperature Trend for Precambrian Life Inferred from Resurrected Proteins. Nature 451:704-707.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Joshua Stern
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules