2010 Annual Science Report
 Montana State University
Reporting | SEP 2009 – AUG 2010
Montana State University
Reporting | SEP 2009 – AUG 2010
Radical SAM Enzyme Functional Diversity and Evolution
Project Summary
The role of radical generating iron-sulfur enzymes in making modification to iron-sulfur motifs in biology are key to the maturation of nitrogen fixing and hydrogen oxidizing enyzme activities. These enzymes act through a mechanism analogous to what has been termed ligand assisted catalysis in discussions of tuning the reactivity of iron sulfur mineral motifs before the advent of life. This strong parallel between biological and abiotic processes provides a basis to better understand the transition from prebiotic chemistry to biochemistry or the transition from the nonliving to the living EArth.
Project Progress
Our progress concerning understanding the chemistry of complex iron-sulfur cluster biosynthesis and iron-sulfur enzyme maturation have brought to light some profound unifying paradigms that link two systems that are of paramount interest to our NAI research, nitrogenases and [FeFe]-hydrogenases. These two enzymes are not evolutionarily related, however, the parallels in the steps they have adapted to synthesize their unique clusters are striking. Both systems start with simple iron-sulfur precursors that are subsequently modified by the activities of radical SAM enzymes and assembled on scaffolds independent of the structural protein. The modification of iron-sulfur motifs by radical S-adenosylmethionine (SAM) enzymes is of particular interest to our NAI project since radical SAM enzymes are iron-sulfur enzymes and all the members of the radical SAM enzyme family act by generating radicals via a common mechanism of reductive cleavage of S-adenosyl methionine to form a radical that is then couple to catalyze hydrogen atom abstraction reactions involving a variety of substrates. For the synthesis of the iron-molydenum cofactor at the active site of Mo-nitrogenases and the H cluster at the active site of [FeFe]-hydrogenases, radical SAM enzymes are involved in key modifications of iron-sulfur motifs and the synthesis and insertion of unique nonprotein ligands that give these specialized complex iron-sulfur clusters their unique reactivity. Conceptually, the involvement of radical chemistry in the modification of iron-sulfur motifs in biology is something we can place in direct context of prebiotic and represent a strong parallel we can exploit to better understand the discontinuity or gap between prebiotic chemistry and biochemistry. The key reactions in FeMo-co and H cluster biosynthesis involve the radical based reaction of simple organics to modify iron-sulfur motifs to support reactivity of central importance in prebiotic chemistry. They represent biology’s version of ligand assisted catalysis. The role of radical SAM enzymes in complex iron-sulfur cluster biochemistry is something that has grown and evolved out of the first generation of results obtained in the first three years of support. The recruitment of radical SAM to these complex biosynthetic pathways is a fascinating independent line of research and one of the perspective we are exploiting to gain further insights into the evolutionary origin of these enzymes. During the previous year of support we have included as a component of our work on complex cofactor biosynthesis, complementary phylogenetic analysis of deduced protein sequences of various biosynthetic enzymes with particular emphasis on the radical SAM enzymes and from this interdisciplinary approach we have gained significant insights how biosynthetic pathways and the cluster themselves evolved.
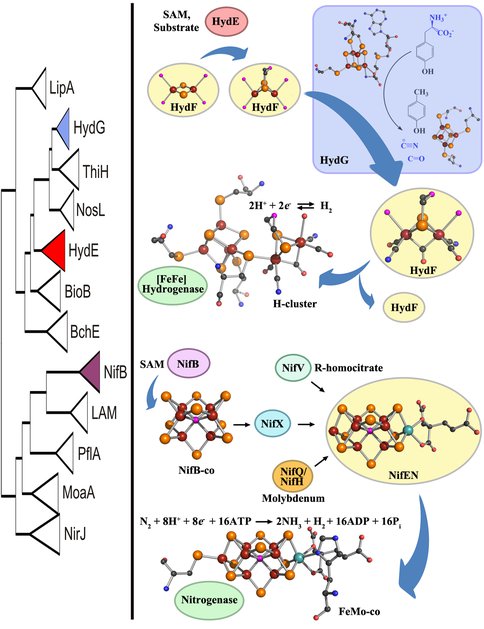
Phylogeny of radical SAM enzymes involved in complex metallocofactor assembly (left). HydE and HydG are involved in H-cluster assembly and NifB is responsible for synthesizing NifB-co, a precursor to FeMo-co. The hypothetical model for H-cluster biosynthesis in [FeFe] hydrogenase maturation (top, right). The evolutionary history of HydE and HydG, relative to other members of the radical SAM protein family, suggests that the emergence of HydE predates the emergence of HydG (left). We propose that this finding supports the hypothesis that HydE performs its chemical modifications prior to HydG during the stepwise synthesis of the H-cluster. HydE utilizes SAM and an unidentified substrate to presumably alkylate a [2Fe-2S] precursor on the scaffold HydF, followed by delivery of CO and CN- via interaction with HydG. In a final step, the 2Fe subcluster on HydF is translocated to the immature structural protein, HydAEFG, presumably through the migration of the 2Fe subcluster through a cationic channel on HydA after which the movement of two loop regions to close the channel. The biochemical model for FeMo-co biosynthesis in nitrogenase maturation (bottom, right). NifB utilizes SAM based radical chemistry to build NifB-co from iron-sulfur cluster precursors. NifB-co is transferred to the scaffold NifEN where molybdenum and homocitrate are introduced to synthesize FeMo-co, which is then transferred to NifDK to yield nitrogenase. Atom colors: maroon (Fe), orange (S), black©, red (O), blue (N), cyan (Mo), pink (unidentified, likely either N or O in the H-cluster and either C, N, or O in FeMo-co).
Publications
-
Boyd, E. S., Anbar, A. D., Miller, S., Hamilton, T. L., Lavin, M., & Peters, J. W. (2011). A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology, 9(3), 221–232. doi:10.1111/j.1472-4669.2011.00278.x
-
Driesener, R. C., Challand, M. R., McGlynn, S. E., Shepard, E. M., Boyd, E. S., Broderick, J. B., … Roach, P. L. (2010). -Hydrogenase Cyanide Ligands Derived From S-Adenosylmethionine-Dependent Cleavage of Tyrosine. Angewandte Chemie International Edition, 49(9), 1687–1690. doi:10.1002/anie.200907047
-
McGlynn, S. E., Boyd, E. S., Shepard, E. M., Lange, R. K., Gerlach, R., Broderick, J. B., & Peters, J. W. (2009). Identification and Characterization of a Novel Member of the Radical AdoMet Enzyme Superfamily and Implications for the Biosynthesis of the Hmd Hydrogenase Active Site Cofactor. Journal of Bacteriology, 192(2), 595–598. doi:10.1128/jb.01125-09
-
McGlynn, S. E., Mulder, D. W., Shepard, E. M., Broderick, J. B., & Peters, J. W. (2009). Hydrogenase cluster biosynthesis: organometallic chemistry nature’s way. Dalton Trans., None(22), 4274. doi:10.1039/b821432h
-
Shepard, E. M., Boyd, E. S., Broderick, J. B., & Peters, J. W. (2011). Biosynthesis of complex iron–sulfur enzymes. Current Opinion in Chemical Biology, 15(2), 319–327. doi:10.1016/j.cbpa.2011.02.012
-
Shepard, E. M., Duffus, B. R., George, S. J., McGlynn, S. E., Challand, M. R., Swanson, K. D., … Broderick, J. B. (2010). -Hydrogenase Maturation: HydG-Catalyzed Synthesis of Carbon Monoxide. Journal of the American Chemical Society, 132(27), 9247–9249. doi:10.1021/ja1012273
-
Soboh, B., Boyd, E. S., Zhao, D., Peters, J. W., & Rubio, L. M. (2010). Substrate specificity and evolutionary implications of a NifDK enzyme carrying NifB-co at its active site. FEBS Letters, 584(8), 1487–1492. doi:10.1016/j.febslet.2010.02.064
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Joan Broderick
Project Investigator
John Peters
Co-Investigator
Eric Boyd
Postdoc
Eric Shepard
Postdoc
Trevor Beard
Doctoral Student
Nicholas Boswell
Doctoral Student
Ben Duffus
Doctoral Student
Rachel Lange
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.1
Sources of prebiotic materials and catalysts
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.3
Origins of energy transduction
Objective 3.4
Origins of cellularity and protobiological systems