2010 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
Georgia Institute of Technology
Reporting | SEP 2009 – AUG 2010
RNA Folding and Assembly
Project Summary
We will characterize the assembly, structure and thermodynamics of the a-PTC by chemical mapping, including hydroxyl radical footprinting1,2 and SHAPE analysis,3 RNase H cleavage, temperature dependent hydrodynamics,4 and computational folding algorithms. In addition we will investigate the effect of freezing aqueous solutions of RNA and DNA molecules on their ability to assemble into larger more complex structures. Freezing nucleic acid solutions concentrates non-water molecules into small liquid pockets in the ice. This enables reactions that can promote the assembly of small segments of nucleic acids into larger complexes.
Project Progress
Structural analysis of the ribosome by our group 1 and by Bokov and Steinberg 2 indicate that the portion of the 23S rRNA comprising the PTC (Peptidyl Transferase Center) is the oldest rRNA fragment in the ribosome’s evolution. We have probed the secondary structure of a proposed 644 nt ancestral rRNA sequence (aPTC) using an RNAse H assay. The assay probed selected segments of the aPTC to determine if they are accessible to DNA oligomer hybridization and RNase H digestion. Segments were targeted based on the expectation that the aPTC folds into the secondary structure observed for this sequence in the 23S ribosomal RNA (see figure 1A). 14 of 15 expected single stranded targets were cleaved over 30% in the assay. The few exceptions may be explained by the structure of these segments in the LSU, which indicate they are buried and may be inaccessible. Surprisingly, 1 of the 3 double-stranded targets, which were not expected to bind to DNA probes and be digested by RNase H, was digested. The target in question, helix 74, was apparently not folded as expected. We are currently examining the influence of 5 truncated rProteins (L2, L3, L4, L15, and L22) as well as solution conditions on the aPTC secondary structure as revealed by the RNase H assay. This work will assist in finding conditions that enable the aPTC to form a structure that functions as a PTC.
Additional studies are being carried out on the assembly of RNA molecules into larger base-paired complexes induced by a small phylogenetic conserved protein called Hfq, or by freezing aqueous solutions. Our recent work indicate Hfq binds to small regulatory RNAs (sRNA) in a 1:1 stoichiometry and ‘melts’ hairpin regions enabling bases to pair with a mRNA target site. Freezing solutions of partially complementary RNAs concentrates them into liquid pockets enabling them to overcome energy barriers and assemble into complexes.
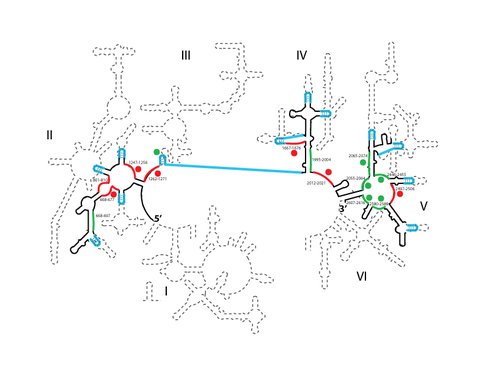
Figure 1A. Original 23S rRNA shown as dotted line. Solid black line connected by blue segments represents the aPTC sequence connected by stem-loop bridges. DNA probes shown in red or green are designated by their target locations in the T. thermophiles 23S rRNA sequence. Red or green circles indicate higher than 30% cleavage by RNase H. One DNA probe targeting all inserted tetraloop sites greatly cleaved one particular tetraloop, as shown with a green circle.
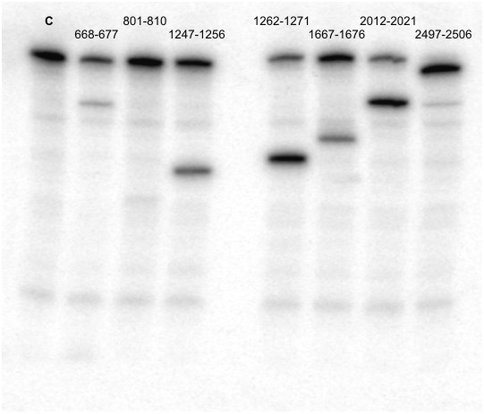
Figure 1B. RNAse H assay results using probes shown in A as red segments on the secondary structure map. Lane C is control lane with uncut aPTC. All probes shown except 801-810 produced 30% or more RNase H degradation.
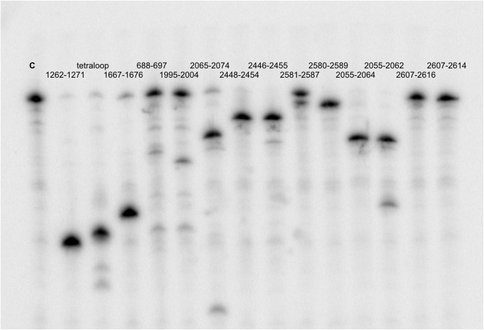
Figure 1C. RNAse H assay results using probes shown in A as green segments on the secondary structure map. Lane C is control lane with uncut aPTC. All probes shown except 688-697 and 1995-2004 produced 30% or more RNase H degradation.
1. Hsiao, C.; Williams, L. D., A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Research 2009, 37 (10), 3134-3142.
2. Bokov, K.; Steinberg, S. V., A hierarchical model for evolution of 23S ribosomal RNA. Nature 2009, 457 (7232), 977-980.
Publications
-
Bokov, K., & Steinberg, S. V. (2009). A hierarchical model for evolution of 23S ribosomal RNA. Nature, 457(7232), 977–980. doi:10.1038/nature07749
-
Hsiao, C., & Williams, L. D. (2009). A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Research, 37(10), 3134–3142. doi:10.1093/nar/gkp119
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Lively Lie
Graduate Student
Taylor Updegrove
Graduate Student
Robin Jacobs
Undergraduate Student
Kanav Jain
Undergraduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 5.3
Biochemical adaptation to extreme environments