2009 Annual Science Report
 Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Reverse-Evolution of an RNA-based RNA Polymerase
Project Summary
The RNA World Hypothesis suggests an RNA molecule is capable both of encoding information and replicating it. In essence, the RNA World Hypothesis predicts an RNA polymerase ribozyme. Since there are no extant RNA-based RNA polymerases, we must instead search the evolutionary fossil record for hints. Our primary goal is to test the hypothesis of Poole that the Small Subunit (SSU) of the ribosome may have evolved from an RNA-dependent RNA polymerase ribozyme.1 We will test the plausibility of an RNA polymerase origin of the SSU by using in vitro reverse evolution; If we can reverse-evolve the SSU into an RNA polymerase, we can demonstrate the a possible evolutionary pathway between a putative primordial ribozyme polymerase and modern ribosomes.
Project Progress
Plausible origin of life scenarios are based upon the RNA World Hypothesis2-4, which suggests that the first self-replicating molecules were made of RNA, and that DNA (optimized for information storage) and proteins (optimized for catalysis) evolved after RNA. The problem of how RNA might have been polymerized by an RNA self-replicase is very much an open question. The simplest route to RNA self-polymerization is not the one that exists in extant protein-based RNA polymerases, so an evolutionary jump would have to have taken place rather early in the history of biochemical evolution. To gain insight into how such an evolutionary event might have taken place, we “reverse-evolved” a ribozyme ligase, which catalyzes the chemical step of the extant RNA polymerization reaction, into a similar ligase, which catalyzes the simpler presumed precursor in chemical evolution.
The L1 ligase is a ribozyme ligase that catalyzes the chemical step of RNA polymerization common to all extant polymerases, namely, condensation of the 3’-OH onto the 5’-alpha-phosphate of a nucleotide triphosphate.5 A more primitive RNA assembly mechanism involving ligation of 2’,3’-cyclic phosphates to the 5’-OH termini of nascent RNA fragments has several evolutionary advantages in terms of more simplified chemistry, as well as abundance of substrates (2’,3’-cyclic phosphates are the natural products of RNA degradation in an aqueous solution). We have, using the methods of in vitro evolution and selection, ‘reverse-evolved’ the L1 ligase into a ligase ribozyme that uses 2’,3’-cyclic phosphate substrates, replacing nucleotide 5’-triphosphate substrates, thus demonstrating a potential evolutionary connection between the two ligases. Remarkably, the new ligase preserves the regiospecific 3’-to-5’ phosphodiester bond formation activity, and does so via a kinetically comparable mechanism. We have in addition crystallized the new 2’,3’-cyclic phosphate ribozyme, and we are attempting to solve its crystal structure from a 3Å resolution X-ray data set. A representative diffraction pattern is provided.
1. Poole, A. M., Jeffares, D. C. & Penny, D. (1998). The Path from the RNA World. J Mol Evol, 46, 1-17.
2. Rich, A. (1962). Horizons in Biochemistry (Kasha, M. & Pullman, B., eds.), pp. 103–126. New York: Academic.
3. Woese, C. R. (1968). The Genetic Code. Harper & Row.
4. Gilbert, W. (1986). Origin of Life – the RNA World. Nature, 319, 618-618.
5. Robertson, M. P. & Scott, W. G. (2007). The Structural Basis of Ribozyme-Catalyzed RNA Assembly. Science, 315, 1549-1553.
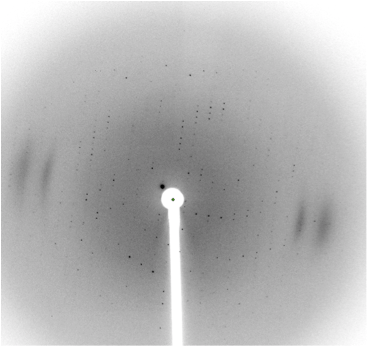
Figure 1. Figure 1. Diffraction from crystals of a the new 2’,3’-cyclic phosphate ribozyme.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Michael Robertson
Postdoc
Alastair Fyfe
Graduate Student
Eric Schultz
Graduate Student
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules