2009 Annual Science Report
 Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Georgia Institute of Technology
Reporting | JUL 2008 – AUG 2009
Experimental Model System - an Ancestral Magnesium-RNA-Peptide Complex
Project Summary
We will develop small model systems in which the interactions of a-rPeptides, Mg2+ ions and a-rRNA can be studied by NMR, X-ray diffraction, calorimetry, molecular dynamics simulations, and other ‘high resolution’ biophysical techniques. Within the large subunit of the extant ribosome, one can observe a tail of ribosomal protein L2 (which we call a-rPeptideL2) that interacts with ribosomal helices rRNA 65 and 66 (which are conserved in a-rRNA), which in turn combines with Mg2+ to form a Mg2+-mc. We will define the smallest a-rRNA and peptide segments (of L2) that are sufficient for assembly of this complex and will characterize the assembly by a variety of experimental and computational methods.
Project Progress
We are developing systems to deconstruct the a-PTC so that we can study the molecular interactions within it. Noller,1 Nierhaus,2 and others showed that extant LSU ribosomal assembly requires only ribosomal proteins L2, L3 and L4, the 23S rRNA, along with inorganic ions such as Mg2+, K+, and Na+. Our model of the a-PTC contains a-rRNA and three a-rPeptides (a-PL2, a-PL3, and a-PL4), with sequences corresponding to the L2, L3, and L4 tails, that drill deeply into the LSU. In addition, the a-PTC contains six highly coordinated Mg2+ ions, which are in the form of three “magnesium microclusters” (Mg2+-μc’s). The sequences and molecular interactions of these ancestral components are conserved over vast evolutionary timescales, predating the last common universal ancestor of life.3-5 Our structural analysis and published biochemical results6 indicated that one of the Mg2+-μc’s plus a-PL2 plus helices 65 and 66 of the a-rRNA (Figure 1) is self-assembling. Our goal here is to recapitulate this a-rRNA/Mg2+/a-rprotein complex in a small model system that will facilitate an integrated experimental and computational investigation of its interactions and assembly.
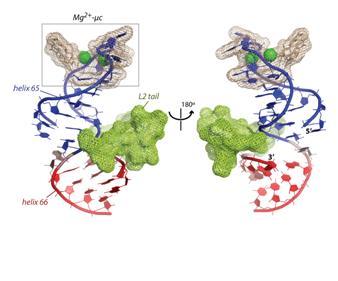
Figure 1. Figure 1 (a) Helices 65 and 66, the L2 tail, and two Mg2+ ions, from structure of T. thermophilus ribosome.
1. Khaitovich, P., Mankin, A. S., Green, R., Lancaster, L. & Noller, H. F. (1999). Characterization of Functionally Active Subribosomal Particles from Thermus Aquaticus. Proc Natl Acad Sci U S A, 96, 85-90.
2. Schulze, H. & Nierhaus, K. H. (1982). Minimal Set of Ribosomal Components for Reconstitution of the Peptidyltransferase Activity. EMBO J, 1, 609-613.
3. Hsiao, C. & Williams, L. D. (2009). A Recurrent Magnesium-Binding Motif Provides a Framework for the Ribosomal Peptidyl Transferase Center. Nucleic Acids Res, 37, 3134-3142.
4. Hsiao, C., Mohan, S., Kalahar, B. K. & Williams, L. D. (2009). Peeling the Onion: Ribosomes Are Ancient Molecular Fossils. Mol Biol Evol, 26, 2415-2425.
5. Hsiao, C., Tannenbaum, M., VanDeusen, H., Hershkovitz, E., Perng, G., Tannenbaum, A. & Williams, L. D. (2008). Complexes of Nucleic Acids with Group I and II Cations. In Nucleic Acid Metal Ion Interactions (Hud, N., ed.), pp. 1-35. The Royal Society of Chemistry, London.
6. Kitahara, K., Kajiura, A., Sato, N. S. & Suzuki, T. (2007). Functional Genetic Selection of Helix 66 in Escherichia Coli 23S rRNA Identified the Eukaryotic-Binding Sequence for Ribosomal Protein L2. Nucleic Acids Res, 35, 4018-4029.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Chiaolong Hsiao
Postdoc
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules

