2009 Annual Science Report
 NASA Ames Research Center
Reporting | JUL 2008 – AUG 2009
NASA Ames Research Center
Reporting | JUL 2008 – AUG 2009
Origins of Functional Proteins and the Early Evolution of Metabolism
Project Summary
The main goal of this project is to identify critical requirements for the emergence of biological complexity in early habitable environments by examining key steps in the origins and early evolution of catalytic functionality and metabolic reaction networks. Using proteins, which are the main catalytic agents in terrestrial organisms, we investigate whether enzymatic activity can arise from an inventory of polymers that have random sequences and that might have existed in habitable environments. We attempt the first demonstration of multiple origins of a single enzymatic function, and investigate experimentally how primordial proteins could evolve through the diversification of their structure and function. Building on this work and on our knowledge of ubiquitous protocellular functions and the constraints of prebiotic chemistry, we conduct computer simulations to elucidate fundamental principles that govern the coupled evolution of early metabolic reactions and their catalysts.
Project Progress
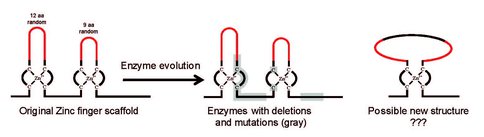
We have recently created de novo an enzyme from a non-catalytic protein structure (scaffold) based on two structural motifs called zinc fingers with randomized loop regions. Amino acid sequence analysis revealed that the new protein very likely adopts a different three-dimensional structure (Fig. 1). To facilitate the analysis of the structure and function of this novel enzyme we have evolved in vitro new variants of it that have higher thermal stability and activity. In addition, the new variants are monomeric and they show a higher cooperativity during thermal unfolding and have higher solubility in water. This allows for structural investigations of the protein using Nuclear Magnetic Resonance (NMR) spectroscopy and X-ray crystallography. These investigations are underway and NMR experiments have already yielded first insights into the structure. The core of the protein appears to be highly structured while the termini are markedly more flexible. Mutants of our protein, in which 13 amino acids at either or both ends were deleted, retain enzymatic activity of at least 10% compared to the full-length enzyme. This indicates that proteins even simpler than the original one remain catalytic.
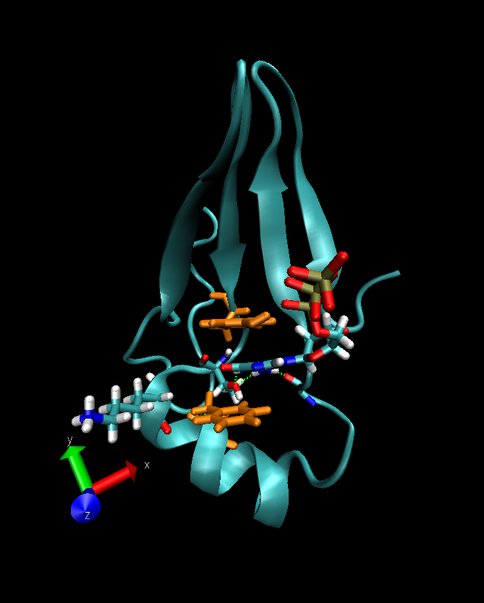
In parallel we initiated computational studies to redesign another simple protein that had been created previously through in vitro evolution (Fig. 2). This protein binds adenosine triphosphate (ATP). The goal is to change its specificity such that, instead, it will bind guanosine triphosphate (GTP). One candidate structure has been already developed. It involves five single-point mutations that allow amino-acid side chains to form three favorable hydrogen bonds with GTP and prevent water from penetrating the core of the protein. Other candidate structures are being designed.
Once completed, these two studies will provide the first evidence that simple proteins that might have existed at the origins of life could evolve and diversify their functions and structures through a small number of point mutations.
Figure 1. De novo enzymes may have adopted a structure different from that of the original protein.
Figure 2. A proposed structure of the redesigned ATP-binding protein that now binds GTP. The guanine part of GTP is located between two aromatic side chains (light brown) and forms three hydrogen bonds with side chains in a loop of the protein (green dashed lines)
Publications
-
Des Marais, D. J., Allamandola, L. J., Benner, S. A., Boss, A. P., Deamer, D., Falkowski, P. G., … Yorke, H. W. (2003). The NASA Astrobiology Roadmap. Astrobiology, 3(2), 219–235. doi:10.1089/153110703769016299
-
Pohorille, A., & Deamer, D. (2009). Self-assembly and function of primitive cell membranes. Research in Microbiology, 160(7), 449–456. doi:10.1016/j.resmic.2009.06.004
-
Wei, C., & Pohorille, A. (2009). Permeation of Membranes by Ribose and Its Diastereomers. Journal of the American Chemical Society, 131(29), 10237–10245. doi:10.1021/ja902531k
- Pohorille, A. (2009, to be published). Contingency vs. determinism in the origin of life. Orig. Life Evol. Biosphere.
- Pohorille, A. (2009, to be published). Emerging properties in the origins of life and Darwinian evolution. Orig. Life Evol. Biosphere.
- Pohorille, A., Wilson, M.A. & Wei, C. (2009, in press). The Earliest Ion Channels. Bioastronomy.
- Pohorille, A., Wilson, M.A. & Wei, C. (2009, in press). The Earliest Ion Channels. In: Meech, K., Keane, J., Mumma, M., Siefert, J. & Werthimer, D. (Eds.). Proceedings of the Bioastronomy 2007 Conference. Astronomical Society of the Pacific.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Burckhard Seelig
Co-Investigator
Michael Wilson
Co-Investigator
James Lake
Collaborator
Milan Mijajlovic
Postdoc
Chenyu Wei
Research Staff
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems