2015 Annual Science Report
 University of Wisconsin
Reporting | JAN 2015 – DEC 2015
University of Wisconsin
Reporting | JAN 2015 – DEC 2015
Project 1C: Analysis of Dissimilatory Iron-Reducing Microbial Communities in Chocolate Pots Hot Spring, Yellowstone National Park
Project Summary
This study represents the first targeted exploration of the active microbial community at source vent of Chocolate Pots hot springs (CP), a warm, circumneutral pH hot spring in Yellowstone National Park. This work was motivated by previous in vitro dissimilatory iron reducing (DIR) incubations of the native microbial community present in the Fe(III) oxide deposits (hereafter referred to as “CP oxides”) near the vent. DIR has the potential to generate distinct signatures of microbial Fe redox metabolism, and identification of the microbial assemblages involved in this metabolism is important for making a concrete linkage between biological metabolism and the generation of geochemical and isotopic biosignatures in relation to redox gradients on Earth and other rocky planets. The central goal of this study was to obtain a phylogenetic and metagenomic characterization of the active acetate-oxidizing DIR community at CP using 13C stable isotope probing (SIP) techniques. CP oxide sediments and spring water were collected from the CP vent source and used to initiate in vitro SIP incubations using labeled (13C) and unlabeled acetate. Incubations targeted the active microbial community which is capable of coupling the oxidation of acetate to DIR. The SIP results allowed us to clearly separate the active acetate-metabolizing microbial community from the rest of the community and identify which organisms native to CP make up this population. The role of some members of this community can be inferred with reasonable confidence from the phylogeny of the OTUs from the amplicon libraries (e.g. Geobacter, Ignavibacteria), and the design of the incubations. The metabolic role of other dominant taxa is less well understood at this point and we are preparing to submit samples for shotgun metagenomic sequencing to address these questions.
Project Progress
Iron (Fe) biogeochemical cycling in circumneutral pH systems is an increasingly important astrobiological target in light of recent discoveries on Mars by Curiosity (Grotzinger et al., 2014). Previous research into microbial ferric iron [Fe(III)] reduction in Yellowstone National Park geothermal springs has focused on high temperature, low pH environments where microbial communities are able to utilize soluble Fe(III) as an electron acceptor for respiration (e.g. Kozubal et al., 2012). Much less attention has been paid to Fe(III)-reducing microbial communities in lower temperature, circumneutral pH environments, where solid phase Fe(III) oxides are the dominant forms of ferric iron.
This study represents the first targeted exploration of the active microbial community at source vent of Chocolate Pots hot springs (CP), a warm (ca. 50°C), circumneutral pH (ca. 5.9-6.7) hot spring in Yellowstone National Park. This work was motivated by previous in vitro dissimilatory iron reducing (DIR) incubations of the native microbial community present in the Fe(III) oxide deposits (hereafter referred to as “CP oxides”) near the vent. DIR has the potential to generate distinct signatures of microbial Fe redox metabolism, and identification of the microbial assemblates involved in this metabolism is important for making a concrete linkage between biological metabolism and the generation of geochemical and isotopic biosignatures in relation to redox gradients on Earth and other rocky planets. The native microbial community was capable of reducing CP oxides, producing substantial amounts of Fe(II) (ca. 60 mmol L-1) in amended (acetate containing) and unamended (no additional electron donor) incubations. The central goal of this study was to obtain a phylogenetic and metagenomic characterization of the active acetate-oxidizing DIR community at CP.
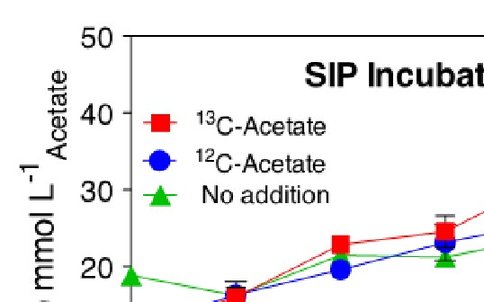
CP oxide sediments and spring water were collected from the CP vent source and used to initiate in vitro DIR incubations and amended, stable isotope probing (SIP) incubations using labeled (13C) and unlabeled acetate. Incubations targeted the active microbial community, which is capable of coupling the oxidation of acetate to DIR. Short duration (10 d) incubations were sampled every 2 d to obtain a profile of reduction activity. Amended samples received 0.5 mM acetate after each sample collection (Figure 1).
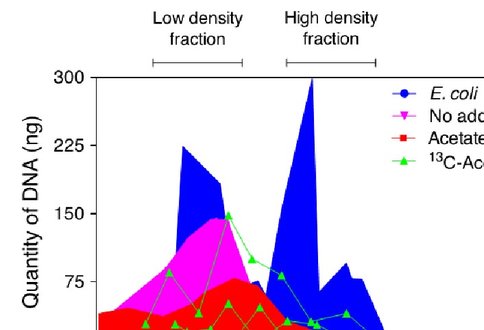
At the end of the experiment, DNA was extracted from the entire contents of the incubation reactors using standard commercial kits. Extraction yielded ca. 1000 ng DNA from each replicate of the of the incubation reactors. Samples were subjected to isopycnic centrifugation to separate low-density DNA from high-density (13C-labeled) DNA, according to previously published techniques (Dunford and Neufeld, 2010). Sixteen gradient fractions were collected for each sample, all of which contained at least some amount of DNA due to the range of densities of DNA corresponding to varying %GC among microorganisms (Panijpan, 1977). Low- and high-density fractions corresponding to light (unlabeled) and heavy (13C-labeled) DNA were identified using a standard of mixed E. coli grown in 12C and 13C media (Figure 2).
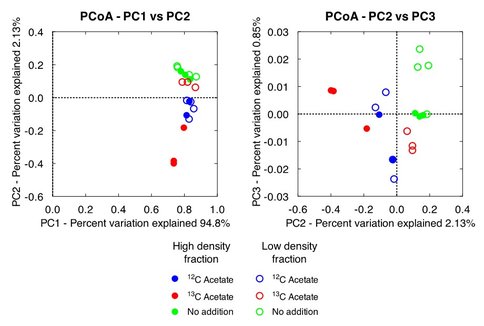
Low- and high-density gradient fractions from each of the three treatments were amplified using Bacterial and Archaeal 16S rRNA gene PCR primers. Amplicons were submitted to the University of Wisconsin Biotech Center for paired-end (2×250 bp) Illumina MiSeq DNA sequencing. Nineteen bacterial amplicon libraries were generated containing between 6000 and 15000 reads at ca. 450 bp in length. Phylogenetic identity of the reads was determined using the QIIME pipeline optimized for processing Illumina datasets (Caporaso et al., 2010; Caporaso et al., 2011). Sequences from the ten most abundant OTUs from each sample were analyzed using NCBI BLASTn search algorithm to obtain additional information about the abundant taxa (Altschul et al., 1990). Comparison of the sequence information was accomplished using the principle coordinate analysis (PCoA) tool as part of the QIIME pipeline.
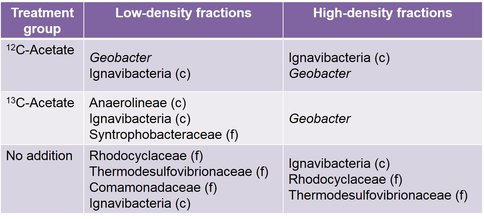
Similar dominant OTUs were present in both high-density and low-density fractions recovered from the unamended samples, including relatives of the families Rhodocyclaceae, Comamonadaceae, Thermodesulfovibrionaceae, and the class Ignavibacteria (Table 1).
While only distantly related to any sequence in the BLAST database (94% similarity), members of the family Rhodocyclaceae have previously been identified as iron-reducers (Cummings et al., 1999). Members of the class Ignavibacteria have also been identified as iron-reducing bacteria (Podosokorskaya et al., 2013). These samples also shared a high amount of similarity to the low-density fraction from the 13C-acetate amended treatments (Figure 3). This is reasonable to expect as this pool of DNA represents the same non-acetate metabolizing community that was identified in the unamended samples. Similar OTUs are present in all acetate-amended samples regardless of whether labeled or unlabeled acetate was utilized, or the DNA came from the high- or low-density fractions. However, a clear dominance of an OTU related to Geobacter hephaestius is seen in the 13C-acetate amended samples. G. hephaestius was isolated from an anoxic rice paddy and has been identified as a sulfur- and iron-reducing bacterium (Janssen, P. H., unpublished, GenBank accession number AY737507.1) This is reasonable to expect as this fraction represents the acetate-metabolizing portion of the microbial community, whereas the unlabeled acetate samples represent what had previously been the dominant members of the microbial community (e.g. OTUs related to Rhodocyclaceae) along side members of the acetate stimulated community (e.g. G. hephaestius). This is made apparent in the principle components analysis of the microbial communities, where the 13C-acetate amended high-density fractions are distinctly different from the rest of the samples (Figure 3). The dominance of a Geobacter-related OTU is not unexpected in the acetate-amended incubations, and has been well documented in previous research (Lovley et al., 2004), however this observation was not made in previous acetate-amended incubations of CP materials.
In summary, this study allowed us to clearly separate the active acetate-metabolizing microbial community from the rest of the community and identify which organisms native to CP make up this population. The role of some members of this community can be inferred with reasonable confidence from the phylogeny of the OTUs from the amplicon libraries (e.g. Geobacter), and the design of the incubations (i.e. iron reducing conditions). The metabolic role of other dominant taxa is less well understood at this point and we are preparing to submit samples for shotgun metagenomic sequencing to begin to address these questions.
References
Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403-410.
Caporaso, J. G., J. Kuczynski, J. Strombaugh, K. Bittinger, F. D. Bushman, and E. K. Costello. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335-336.
Caporaso, J. G., C. L. Lauber, W. A. Walters, D. Berg-Lyons, C. A. Lozupone, P. J. Turnbaugh et al. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108 Suppl 1:4516-4522.
Cummings, D. E., F. Caccavo Jr., S. Spring, and R. F. Rosenzweig. 1999. Ferribacterium limneticum, gen. nov., sp. nov., an Fe(III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch Microbiol 171:183-188.
Dunford, E. A., and J. D. Neufeld. 2010. DNA stable-isotope probing (DNA-SIP). J Vis Exp.
Grotzinger, J. P., D. Y. Sumner, L. C. Kah, K. Stack, S. Gupta, L. Edgar et al. 2014. A habitable fluvio-lacustrine environment at Yellowknife Bay, Gale Crater, Mars. Science 343:DOI: 10.1126/science.1242777.
Kozubal, M. A., W. P. Inskeep, and R. E. Macur. 2012. Geomicrobiology of iron oxide mats from acidic geothermal springs in Yellowstone National Park: Microbial community structure, isolation of novel species and mechanisms of iron oxidation. Front. Microbiol. 3:109.
Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) Reduction. Adv Microb Physiol 49:219-286.
Panijpan, B. 1977. The Buoyant Density of DNA and the G+C Content. J Chem Educ 54:172-173.
Podosokorskaya, O. A., V. V. Kadnikov, S. N. Gavrilov, A. V. Mardanov, A. Y. Merkel, O. V. Karnachuk et al. 2013. Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae. Environ Microbiol 15:1759-1771.
Publications
-
Fortney, N. W., He, S., Converse, B. J., Beard, B. L., Johnson, C. M., Boyd, E. S., & Roden, E. E. (2016). Microbial Fe(III) oxide reduction potential in Chocolate Pots hot spring, Yellowstone National Park. Geobiology, 14(3), 255–275. doi:10.1111/gbi.12173
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Eric Boyd
Co-Investigator
-
RELATED OBJECTIVES:
Objective 2.1
Mars exploration.
Objective 4.1
Earth's early biosphere.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.3
Biochemical adaptation to extreme environments