2015 Annual Science Report
 University of Montana, Missoula
Reporting | JAN 2015 – DEC 2015
University of Montana, Missoula
Reporting | JAN 2015 – DEC 2015
Project 7: Error Rate and the Origin and Early Evolution of Life
Project Summary
Our project investigates the evolutionary relationship between rates of genetic mutation and genetic recombination. It addresses very general questions about the stability of heredity and the implications of that stability for adaptation and persistence of organisms. Such questions are likely to apply wherever and whenever life evolves. In prior theory work we have shown that the mutation rate of a population will tend towards ever-higher values in the absence of genetic recombination. Because mutation is the ultimate source of the variation required for the evolution of a population, it might be thought that a high mutation rate would enable more rapid evolutionary adaptation. We and others have shown, however, that too high a mutation rate can cause extinction of a population. Because early life probably had very high mutation rates, early life would have been at considerable risk of evolving a lethal mutation rate. This should have produced strong pressure for genetic recombination to evolve. In our project we are using experimental evolution, analytical theory, and computer simulations to test the effect that recombination has on mutation rate evolution, the effect that high mutation rates have on population adaptation and persistence, and the effect of mutation on the evolution of cooperation among life forms.
Project Progress
Significant accomplishments have been made on four distinct sub-projects addressing the main themes of our NAI project. Figures associated with these projects are referenced by number and provided in section 5, below.
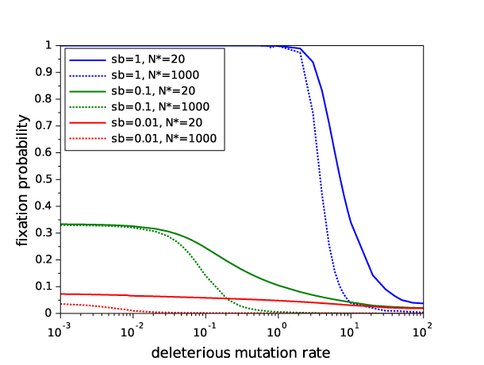
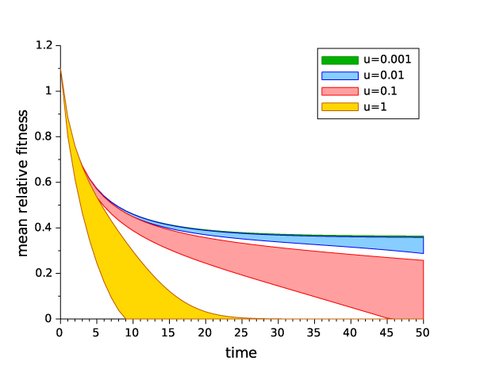
1. A theoretical study of the effects of deleterious mutations on the fate of beneficial mutations in evolving asexual populations: In preliminary computer simulations, we observed that the fixation probability of beneficial mutations in asexual populations is decreased by high deleterious mutation rates; in particular, this is true even in the complete absence of clonal interference from alternative beneficial mutations and background selection against deleterious mutations arising in genomes without the beneficial mutation. We hypothesized that the lower probability of fixation of a beneficial lineage in this situation can be attributed to contamination of the lineage of a beneficial mutation by newly arising deleterious mutations: specifically, because the lineage size of a new beneficial mutation is extremely small, the general process of genome deterioration called Muller’s ratchet operates at a much faster rate in this small lineage than in the rest of the population. In a manuscript that is about to be submitted, we present results of analytical modeling and further computer simulations that support this hypothesis and show how lineage contamination affects fixation probabilities, dynamics of differential load (relative fitness dynamics), sojourn dynamics, and fitness effects of surviving beneficial mutations.
Significantly, we show that lineage contamination alone is the factor that can cause a population to stop substituting beneficial mutations and decline in fitness toward extinction. We relate our results to the classical “error threshold” idea and discuss potential experimental approaches related to the theory developed. The manuscript, which is entitled “Dynamics and fate of beneficial mutations under lineage contamination by linked deleterious mutations”, has as co-authors NAI Co-I Gerrish, Lead Sniegowski, Collaborator Singh, and international non-NAI collaborator Dr. Sophie Pénisson of Université Paris-Est, France.
2) Inferring the distribution of fitness effects of newly arising mutations: Classical population and quantitative genetic approaches model evolution based on the standing allelic variation in populations. The standing allelic variation itself, however, is ultimately dependent on input from mutation, which supplies the raw material with which natural selection works. Understanding the distribution of fitness effects (DFE) of new mutations is thus a central concern of evolutionary genetics. Despite some recent progress in understanding the DFE in well defined systems, large gaps remain. In particular, no study to date has characterised the DFE in a population over time, as it evolves. In this project, we have developed a powerful new mathematical framework for analyzing the DFE based on time-series samples of an evolving experimental population. The results will allow the first look at the changing landscape of mutations affecting fitness as a population evolves. Collaborators and co-authors on this work include NAI Co-I Gerrish, Lead Sniegowski, and Collaborator Singh; submission of a theory paper is anticipated in spring 2016, with empirical work to follow.
3. Mutation accumulation and mutation rate evolution in populations with extraordinarily high genomic mutation rates: As noted above, our previous work has indicated that populations without recombination are likely to evolve very high mutation rates. This then raises questions about the fate of such populations: Will they go extinct? Will they eventually evolve decreased mutation rates and persist? These questions are likely to apply to life forms wherever they may originate and evolve, as they bear on the fundamental relationship between the random origin of hereditary variation and adaptation through natural selection. In Year 1 of our NAI project, NAI Collaborator Singh carried out an experiment in which she used small population size to allow accumulation of mutations in experiment populations of E. coli engineered by our lab to have mutation rates 5,000 higher than wild type. Of the 60 populations propagated in this work, approximately half went extinct; of the remaining populations, about half show evidence of reduced genomic mutation rates. Along with NAI collaborator Vaughn Cooper at the University of Pittsburgh, we are now obtaining genome sequences from these populations in order to analyze the nature and spectrum of accumulated mutations and to identify those changes that lead to decreased mutation rates. Submission of multiple papers from this work is anticipated during 2016, with coauthors including NAI Project Collaborator Singh, Lead Sniegowski, and Pittsburgh NAI Lead Cooper.
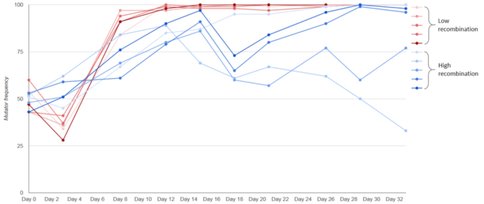
4. Recombination and the dynamics of mutator alleles: A fundamental goal of our NAI project is to understand, both theoretically and empirically, the relationship between recombination and mutation rate. NAI Collaborator Galeota-Sprung has made significant progress on this aspect of our project in Year 1, carrying out a series of studies on the fate of mutator alleles (which increase the genomic mutation rate) in experimental E. coli populations engineered so that they will exhibit high vs. low genomic recombination via conjugative transfer of DNA. These experiments have examined whether recombination can impede or even prevent the rise to fixation of a mutator allele, thus saving populations from the tendency to evolve ever-higher mutation rates. The results have been striking, as shown in Fig. 3: mutators rise quickly toward fixation in low-recombination populations via a process known as genetic hitchhiking, but high recombination rate impedes this process.
A manuscript describing these and other results, as well as related computer simulations, is currently in preparation. Its authors include NAI Project Collaborator Galeota-Sprung, Lead Sniegowski, and non-NAI collaborator Dr. Eugene Raynes, who is at Brown University. Notably, NAI Collaborator Galeota-Sprung is applying for an Astrobiology Early Career Collaboration Award to collaborate with University of Pittsburgh NAI Lead Vaughn Cooper on sequence analyses of these populations. Dr. Cooper will be a coauthor on future papers exploring sequence evolution in the system.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Philip Gerrish
Co-Investigator
Mitra Eghbal
Collaborator
Benjamin Galeota-Sprung
Collaborator
Tanya Singh
Collaborator
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.
Objective 5.2
Co-evolution of microbial communities
Objective 6.2
Adaptation and evolution of life beyond Earth