2015 Annual Science Report
 University of Montana, Missoula
Reporting | JAN 2015 – DEC 2015
University of Montana, Missoula
Reporting | JAN 2015 – DEC 2015
Project 3: Consequences of recA Duplication for Recombination, Genome Stability and Fitness
Project Summary
Homologous recombination (HR) – the exchange of genetic information between similar DNA molecules – is an ancient process that is central to the emergence of biological complexity, diversity and stability. Yet, it must be tightly regulated, as it is likewise an important source of destabilizing genomic rearrangements. Despite the importance of HR, we still have a poor understanding of the balance of these creative, stabilizing and destabilizing contributions to organismal fitness, complexity and genome evolution. We are using the extraordinary genome evolutionary dynamics and duplicated copies of the HR gene recA in the cyanobacterium Acaryochloris as a model to gain novel insights on the fitness consequences that emerge from the interplay between HR-mediated maintenance of genome stability, selectively favored gene duplications and non-adaptive genomic rearrangements.
Project Progress
We have made substantial progress during the first year of the award period, detailed below.
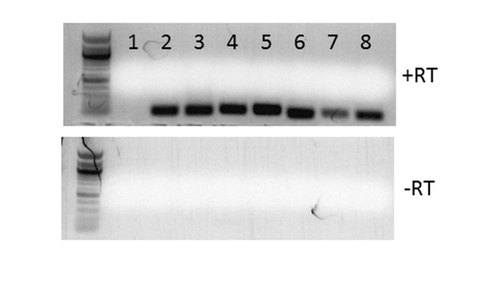
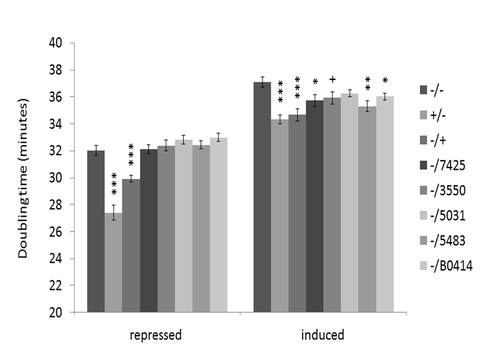
(1) Development of an E. coli model for the characterization of Acaryochloris RecAs. A major question regarding the molecular evolutionary origins of biological diversity is what happens to a generalist enzyme like RecA upon duplication. We have developed tools for investigating the functional divergence of Acaryochloris RecAs using the genetic model bacterium E. coli. We constructed plasmids to specifically induce the expression of recA genes from Acaryochloris in E. coli in the presence of the sugar rhamnose. We can detect both RNA transcripts (Fig. 1) and protein for Acaryochloris RecAs, indicating that the genes are successfully expressed. We next tested whether the Acaryochloris genes could complement (i.e., rescue) a number of defects observed for an E. coli mutant that lacks its own copy of recA. We observed that no Acaryochloris recA could complement the well-characterized sensitivity of the mutant to ultraviolet radiation or mutagens such as mitomycin C; however, this appears to be a general property of cyanobacterial RecAs that we tested. Interestingly, they could complement the general growth defect of the mutant, which requires recombinational repair activity, to different degrees (Fig. 2), indicating that: (1) Acaryochloris recAs have indeed functionally diverged; and (2) all copies have recombinase activity, consistent with our model that multiple recA copies contribute to the extraordinary genome instability of Acaryochloris through enhanced recombination.
(2) Developing tools to study the function of purified Acaryochloris RecAs. We are following up on this evidence for functional divergence with a study of purified RecA proteins, which will enable finer dissection of the functional divergence of Acaryochloris RecAs. To this end, we have initiated a collaboration with Michael Cox of the University of Wisconsin-Madison, a world leader on RecA biochemistry. We are currently optimizing conditions for the expression of Acaryochloris RecAs in E. coli to obtain high yields of purified protein. In year two of the award, Emiko Sano, a NAI postdoctoral researcher, will visit the Cox lab to receive training in both RecA purification and different biochemical assays of RecA function.
(3) An Acaryochloris genetic system. One goal of the project is to directly test the impact of recA dosage on genome evolution and fitness by manipulating the number of recA copies in Acaryochloris cells. We will do so by deleting specific gene copies either singly or in combination. We have cloned in-frame deletions of Acaryochloris recA genes into different conjugative plasmids for introduction into Acaryochloris cells. We are currently attempting to integrate these deletions into the Acaryochloris genome by both conjugation and electroporation approaches.
(4) Communicating our accomplishments. We are preparing a manuscript based on our results from the E. coli model. In addition, we have presented our work at several meetings during year one of the award. These include:
E. B. Sano, A. L. Gallagher and S. R. Miller. Exceptional gene duplication dynamics in the cyanobacterium Acaryochloris: A possible role for RecA paralogs in adaptation and genome instability. NASA AbSciCon, Exploring the Effects of Stress on Microbial Mutation Rates and Survival Strategies. Chicago, IL. June 2015.
S. R. Miller. Evolutionary consequences of recA duplication in the cyanobacterium Acaryochloris. Invited talk, Microbial Population Biology Gordon Research Conference. Andover, NH. July 2015.
E. B. Sano, A. L. Gallagher and S. R. Miller. Exceptional gene duplication dynamics in the cyanobacterium Acaryochloris: A possible role for RecA paralogs in adaptation and genome instability. Satellite meeting of the Society for Molecular Biology and Evolution (SMBE) on Mechanisms of Protein Evolution. Denver, CO. November 2015.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Amy Gallagher
Co-Investigator
Emiko Sano
Co-Investigator
Michael Cox
Collaborator
-
RELATED OBJECTIVES:
Objective 4.2
Production of complex life.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 6.2
Adaptation and evolution of life beyond Earth