2015 Annual Science Report
 University of Montana, Missoula
Reporting | JAN 2015 – DEC 2015
University of Montana, Missoula
Reporting | JAN 2015 – DEC 2015
Project 1: From Generalist to Specialists (Or Not): A Case Study in Enzyme Evolution
Project Summary
Metabolic enzymes, although prodigious catalysts, are not perfectly specific for their physiological substrates. They typically possess secondary activities as a consequence of the assemblage of highly reactive functional groups, metal ions and cofactors in their active sites. Secondary activities that are physiologically irrelevant, either because they are too inefficient to contribute to fitness or because the enzyme never encounters the substrate, are termed promiscuous activities.
Promiscuous activities are important from an evolutionary standpoint because they provide a reservoir of catalytic potential within a proteome that can be drawn upon when the environment changes. A promiscuous activity may become important for fitness when a new source of carbon, nitrogen or phosphorous appears in the environment, or when a previously available compound, such as an amino acid or cofactor, becomes unavailable. A promiscuous activity may also become critical when the organism is exposed to a novel toxin, such as an antibiotic or pesticide.
A newly recruited promiscuous activity is unlikely to be the optimal solution to an environmental challenge or opportunity. In this project, we are using a model system in E. coli to characterize the genetic changes by which a gene encoding an enzyme whose promiscuous activity has become essential for growth duplicates and diverges to encode a pair of genes encoding efficient specialist enzymes. This work will provide a better understanding of the process by which large superfamilies of enzymes have diverged from generalist enzymes in the last universal common ancestor.
Project Progress
When microbes are faced with an environmental challenge or opportunity, pre-existing enzymes with promiscuous secondary activities can be recruited to provide newly important catalytic functions. Mutations that increase the efficiency of a new activity often compromise the original activity, resulting in an inefficient bifunctional enzyme. Under such conditions, duplication/amplification of the gene encoding the “weak-link” enzyme can increase its expression and consequently increase fitness. Mutations that occur in various copies of the gene can improve the level of one or the other of the essential activities. As improved enzymes evolve, the number of gene copies will decrease, leading ultimately to a pair of genes encoding specialized enzymes. The existence of large superfamilies of enzymes attests to the importance of this process. However, the dynamics of the process itself are poorly understood.
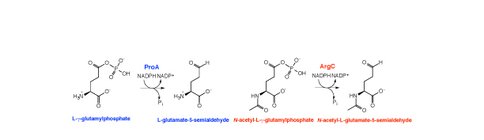
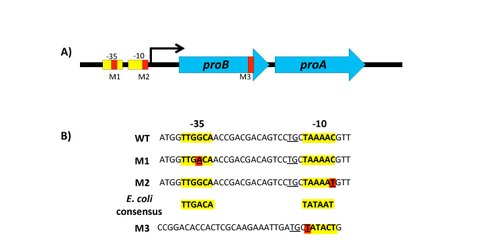
We are examining the process of gene duplication/amplification and divergence in the case of E. coli E383A ProA, which can serve the functions of both ProA (essential for proline biosynthesis) and ArgC (essential for arginine biosynthesis), albeit poorly (see Fig. 1). Since both activities are essential for growth on glucose, E383A ProA is the ‘weak-link” enzyme that limits growth rate in a strain that lacks argC. We have previously shown that growth rate can be increased by massive gene amplification, by point mutations in the promoter, and by a synonymous mutation in the upstream gene in the operon that creates a new transcriptional start site for the gene encoding E383A ProA (see Fig. 2). The goal of our NAI project is to follow the appearance of mutations in various alleles of an amplified array and to characterize the encoded enzymes throughout the process of divergence to two specialized enzymes.
During the past year, we have made progress on this study in several directions.
1) Dr. Juhan Kim and graduate student Andrew Morgenthaler have shown that strains promoter mutations or the synonymous mutation that creates a new transcriptional start site do not preculde gene amplification, although fewer copies are accumulated compared to the parental strain (which lacks argC and carried the allele encoding E383A ProA).
2) Dr. Johannes Rudolph has developed an improved assay for the two activities provided by E383A ProA. While the assay for ArgC activity is straightforward, the assay for ProA activity is not. The substrate for ProA, gamma-glutamyl phosphate, is unstable, and is channeled to the active site of ProA from the active site of glutamyl kinase (ProB). Dr. Rudolph has purified ProB and developed conditions for assaying ProA activity in the presence of the ProB-gamma-glutamyl phosphate complex. The availability of this assay sets the stage for assays of variant enzymes that arise in amplified arrays as the gene encoding proA diverges under selection for faster growth on glucose.
3) Andrew Morgenthaler is engineering a strain of E. coli that carries i) a deletion of argC; ii) the allele encoding E383A ProA; iii) a gene encoding yellow fluorescent protein (YFP) immediately next to proA to enable detection of segmental amplifications; iv) introduced sites for cleavage by restriction enzymes to enable recovery and sequencing of individual proA* alleles. This strain will allow us to follow amplification and de-amplification based on the fluorescence of YFP and to identify the mutations present in each allele of an amplified array. He is also building turbidostats so that we can carry out up to 9 parallel experimental evolution experiments.
4) Undergraduate JohnCarlo Kristofich is examining the mechanisms by which fitness can be improved in a different bacterium (Salmonella enterica) when growth is limited by the inefficiency of E385A ProA (the variant corresponding to E383A ProA in E. coli). In S. enterica, in contrast to E. coli, duplication/amplification of the region surrounding proA does not occur. Rather, an initial promoter mutation increases the expression of the gene encoding E385A ProA. Subsequently, additional point mutations occur upstream of the proA gene and within the proA gene itself. This work demonstrates that the evolutionary trajectory for improvement of fitness when growth rate is limited by an inefficient bifunctional enzyme is considerably different in E. coli and S. enterica.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
John Carlo Kristofich
Co-Investigator
Andrew Morgenthaler
Co-Investigator
Johannes Rudolph
Co-Investigator
Juhan Kim
Collaborator
-
RELATED OBJECTIVES:
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.2
Adaptation and evolution of life beyond Earth