2015 Annual Science Report
 University of Illinois at Urbana-Champaign
Reporting | JAN 2015 – DEC 2015
University of Illinois at Urbana-Champaign
Reporting | JAN 2015 – DEC 2015
Project 8: The Evolution of the Eukaryote-Archaea Common Ancestor
Project Summary
The goals of our lab with respect to the NAI project are to describe early evolutionary via genomic and cellular comparisons of diverse eukaryotes to diverse archaea. We are interested in comparing genomes from diverse free-living eukaryotes to investigate the origins and evolution of eukaryotic complexity. Evolutionary reconstructions of early eukaryotes are challenged by a lack of sufficient taxonomic sampling. Few genomes of free-living microbial eukaryotes are sequenced, despite their critical importance in ecology, evolution, and basic cellular biology. The real challenge to protistan genomics is actually quite mundane; it concerns the lack of available and cultivatable free-living protists (mainly heterotrophs) in the laboratory. Yet, a better understanding of the genomic content diverse eukaryotes facilitates the evolutionary analysis of archaeal genomes. To address these issues of poor taxonomic sampling of eukaryotic genomes, my lab has developed a molecular method to separate eukaryotic DNA from bacterial DNA. We have demonstrated conclusively that we can separate eukaryotic chromatin from a mixture of eukaryotic and bacterial genomic DNA. This method will be widely applicable to the study of protistan genomics. Currently, our lab is in the process of assembling and annotating ten eukaryotic genomes from my lab’s culture collection of over 100 amoeboid protists from diverse phylogenetic groups. Many of these amoeba represent novel phyla-level lineages of eukaryotes.
One amoebal genome is form is Nuclearia sp., which is an amoeboid protist closely and a member of a primary “supergroup” of eukaryotes – the Optisthokonts. This supergroup includes all animals, fungi, and several types of unicellular or colonial protists including choanoflagllates. Thus, genomic analyses of Nuclearia will inform the evolution of complexity and multicelllularity in both Fungi and Animals.
Project Progress
The goals of our lab with respect to the NAI project are to describe early evolutionary via genomic and cellular comparisons of diverse eukaryotes to diverse archaea. My lab is interested in early eukaryotic evolution, and has been involved in the recent sequencing and analysis of the genomes of the free-living amoeboflagellate Naegleria gruberi.
Few genomes of free-living microbial eukaryotes are sequenced, despite their critical importance in ecology, evolution, and basic cellular biology. The real challenge to protistan genomics is actually quite mundane; it concerns the lack of available and cultivatible free-living protists (mainly heterotrophs) in the laboratory. As with bacteria, most free-living protistan taxa are difficult to obtain in pure culture, and virtually all deposited in culture collections as mixed consortia with bacteria. Evolutionary reconstructions of early eukaryotes are challenged by a lack of sufficient taxonomic sampling. There are very few genomes of free-living eukaryotes, because they are grown with bacteria as food, and are not in pure culture.
Eukaryotic genomes are considerably larger and more complex that bacterial or archaeal genomes. We estimate each microbial eukaryotic genome to be over 50 Mb in size (comparable bacterial genomes are closer to 5 Mb). Yet, a better understanding of the genomic content diverse eukaryotes facilitates the evolutionary analysis of archaeal genomes.
However, my lab has developed a molecular method to separate eukaryotic DNA from bacterial DNA (see gel quantification of separation, below). We have demonstrated conclusively that we can separate eukaryotic chromatin from a mixture of eukaryotic and bacterial genomic DNA. This method will be widely applicable to the study of protistan genomics.
Further this method has allowed us to prepare genomic DNA libraries for complete genome sequencing from ten free-living eukaryotic amoebae that we isolated and have characterized from anoxic sediments (genomic DNA of five of 10 are shown in Figure 1: P5, L3A, SW, PL2, and SM) Each of these isolates represents a novel group of eukaryotes – groups that are not related to known phyla of eukaryotes. We are in the process of assembling and annotating these eukaryotic “metagenomes” and developoing computational methods to annotate eukaryotic contigs from mixed community metagenomic data.
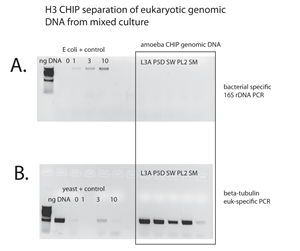
This is a considerable and noteworthy achievement for such a small collaborative working group (e.g., myself and one graduate student). Our comparative genomic analysis will provide insight into the early evolution of eukaryotes, and genes shared with archaea. Currently we are comparing these genomes to other eukaryotes to assess early features of eukaryotes. This is essential concerning the recent publication of archaeal metagenomic data suggesting that archaea have eukaryotic proteins such as actin-related proteins. We expect to submit a manuscript about one or more of these novel eukaryotic genomes in the first quarter of 2016.
Theories of early eukaryotic evolution involve symbioses of bacteria with archaea, yet there are no contemporary examples of these associations. These notions also include a transition in eukaryotes from anaerobic archaea to aerobic eukaryotes via bacterial endosymbiosis.
With respect to staff, I have had support for one graduate student as a collaborator with the U-Illinois NAI. Marissa Hirst’s graduate work was supported by my subcontract (2013-2014). Marissa developed imaging methods for whole cell rRNA FISH of large bacteria that we applied and adapted to isolating and analyzing archaeal symbionts of anaerobic rumen ciliates using FACS. Using metagenomic analysis and fluorescent imaging of sorted anaerobic ciliates, we demonstrated that the methanogenic archaea have genomes that are suggestive of adaptions to life inside a eukaryote cell as an endosymbiont. This is the first genomic analysis of a methanogenic archaeal endosymbiont of a eukaryote.
Publications
-
Prokopenko, M. G., Hirst, M. B., De Brabandere, L., Lawrence, D. J. P., Berelson, W. M., Granger, J., … Sigman, D. M. (2013). Nitrogen losses in anoxic marine sediments driven by Thioploca–anammox bacterial consortia. Nature, 500(7461), 194–198. doi:10.1038/nature12365
- On road to archaeal symbiosis: genomic reduction in novel Methanobrevibacter sp. associated with rumen ciliates. (2015) Hirst, M.B., Karberg, K. and Dawson, S.C. (In Review)
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Isaac Cann
Co-Investigator
Gary Olsen
Co-Investigator
Rachel Whitaker
Co-Investigator
Matthew Wright
Co-Investigator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.2
Production of complex life.
Objective 6.2
Adaptation and evolution of life beyond Earth
