2015 Annual Science Report
 University of Illinois at Urbana-Champaign
Reporting | JAN 2015 – DEC 2015
University of Illinois at Urbana-Champaign
Reporting | JAN 2015 – DEC 2015
Project 4: Experiment on Darwinian Transition
Project Summary
Carl Woese proposed that life started as semi-autonomous subcellular forms named progenotes. The progenotes lacked cell membranes and readily exchanged information, suggesting that aspects of information processing had already been developed. Woese further hypothesized that certain early life processes crossed a Darwinian threshold, where incorporation of new components of a processes was not tolerated. We aim at determining whether translation, transcription, and replication have crossed the Darwinian threshold. To determine whether DNA replication has crossed the Darwinian Threshold, interchangeability of the DNA replication processivity factor known as the sliding clamp is being examined. It is only in the presence of the sliding clamp that DNA polymerases in extant organisms can gain the speed required to replicate their genomes. In Bacteria, the sliding clamp is the -subunit of Pol-III and in Archaea and Eukarya the functional homolog is proliferating cell nuclear antigen (PCNA). We have, therefore, expressed and purified a sliding clamp from each of the three domains of life (E. coli -subunit, M. acetivorans PCNA, and human PCNA). Sliding clamps are loaded in a clamp loader dependent manner; therefore, we have cloned, expressed and purified an archaeal clamp loader from M. acetivorans. Our next step is to determine whether an archaeal clamp loader can interact with each of the sliding clamps from the three domains of life and whether any of the interactions leads to loading of the sliding clamps onto DNA to orchestrate processive DNA synthesis.
Project Progress
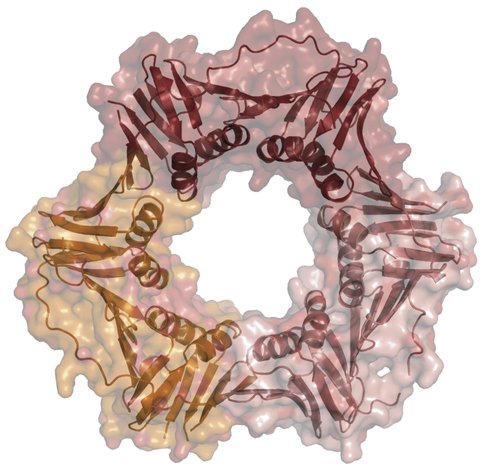
Our experimental work focuses mostly on two proteins that are central to the replication of the genome in all three forms of life. These proteins are namely the sliding clamp (Fig. 1), a donut shaped protein, and a clamp loader, a five sub-unit protein complex, that opens the sliding clamp and loads it onto the DNA. Once the sliding clamp is loaded onto the DNA, the DNA polymerase (enzyme that copies the genome) binds to the sliding clamp and travels at the rapid speed by which the genome can be duplicated in a rapid manner for transfer to daughter cells (progeny). Note that in the absence of the sliding clamp, a DNA polymerase such as Pol-III which replicates the E. coli genome will synthesize DNA at 10 nucleotides per second, whereas when bound to its sliding clamp, it will synthesize 700 – 1000 nucleotides per second. Thus, these proteins are central to extant cells evolving large genomes. There are several steps required for the sliding clamp to be loaded onto the DNA to perform its function. The first step requires the clamp loader to bind to the clamp in the presence of ATP. In our experiments, we are using Methanosarcina acetivorans clamp loader (i.e., an archaeal clamp loader) to load 3 sliding clamps from the three domains of life. The three sliding clamps are MacPCNA, the beta-subunit, and HsaPCNA, which represent the sliding clamps from M. acetivorans (Archaea), E. coli (Bacteria) and human (Eucarya), respectively. The first step required for the clamp loader to load the sliding clamp is interactions of the two proteins in the presence of ATP. We have used size exclusion chromatography for this analysis, and the results clearly show that the clamp loader of the archaeon M. acetivorans does not interact with the E. coli beta-subunit. This finding suggests that the _E. coli sliding clamp cannot substitute for the archaeal sliding clamp. In other words, if one considers this central component within the replication machinery from the archaeal and bacterial perspective, the replication machinery has crossed the Darwinian threshold. However, for such a significant hypothesis underlying evolution of life on this planet, it is important that we use several approaches and proteins to confirm this observation, and currently we are carrying out experiments both in vitro and in vivo to ascertain our observation.
We are also carrying out experiments using the DNA replication machinery, from both genetic and biochemical approaches, to determine the nature of the common ancestor of the Archaea/Eukarya sister domains before they split into two different domains. A critical component of the DNA replication machinery is a protein that marks where DNA replication should initiate in the genome. This protein is functionally conserved universally and is commonly called the origin recognition protein/complex. Our experimental work based on Methanosarcina acetivorans demonstrates that before the Archaea and Eukarya diverged, their common ancestor had already developed an origin recognition protein that is different from the one currently present in the domain Bacteria. This finding together with other biochemical, structural and bioinformatics analyses allow us to conclude that the common ancestor of the Archaea/Eukarya sister domains had already evolved a large genome with a sophisticated machinery to replicate the genome for transfer to progeny. We intend to further test if the simpler origin recognition protein from the archaeon M. acetivorans can replace the origin recognition protein or DnaA in the bacterium E. coli. This experiment will also be an important step in our effort to determine whether the replication machinery has crossed the Darwinian Threshold. A critical finding from this work shows that when the archaeal homolog of the protein that marks where replication should start is shut down, the cells grow in size, presumably in preparation to divide. However, because replication never starts the cell only grows in size (Fig. 2) and never divides.
Our studies on single-stranded DNA binding proteins, which likely co-evolved with DNA as the molecule that stores genetic information, suggests that a diverse group of this protein was invented in life, a scenario that captures Woese’s progenote state. We have further discovered that while bacteria and eukaryotes seems to contain single-stranded DNA (ssDNA) binding proteins with common structural fold, some archaea such as M. acetivorans contain ssDNA binding proteins that are a mixture of both the bacterial and eukaryotic structural fold. We continue to analyze these proteins to gain insight into their potential nature and whether they truly reflect entities from Woese’s progenote state from which extant life or cellular is predicted to have evolved.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Zaida Luthey-Schulten
Co-Investigator
Carla S.P. Baptista
Collaborator
Thomas Cyberski
Collaborator
Jaigeeth Deveryshetty
Collaborator
Jacob Farris
Collaborator
Matthew Heffernan
Collaborator
Yoshizumi Ishino
Collaborator
Alexander Nemeh
Collaborator
Miyako Shiraishi
Collaborator
Kyle Tegtmayer
Collaborator
Noopur Walia
Collaborator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.2
Production of complex life.