2015 Annual Science Report
 Massachusetts Institute of Technology
Reporting | JAN 2015 – DEC 2015
Massachusetts Institute of Technology
Reporting | JAN 2015 – DEC 2015
Early Animals: Sensory Systems and Combinatorial Codes
Project Summary
Understanding the evolution of integrated sensory organs—such as the eyes, ears and nose that develop in concert on our heads—is fundamental to understanding animal complexity. These are the features that permit movement and the environmental responses that characterize animals. We examine understudied early branches of the animal family tree, with a focus on the jellyfish Aurelia, to understand how the genetic regulation of sensory organs is conserved in some cases and evolves in others. Comparison of developmental regulation reveals how similar gene networks can be differentially modified and deployed, permitting the evolution of complex sensory systems. Jellyfish provide an ideal study system for the examination of the evolution of such sensory systems in animal evolution, as they are the most basal branch of the animal tree with multiple sensory modes, and these develop at multiple stages in a complex life history. This provides us the ability to compare and contrast within the broader cnidarian group to which jellyfish belong, and to the bilaterians, the broad group containing humans and most other animals. The application of genomic methods greatly enhances our ability to pursue these questions.
Project Progress
Aurelia, the moon jellyfish provides an excellent model for the evolution of complex sensory systems. It possesses multiple sense organs that operate in different sensory modalities in a complex life history. Cnidaria are the most basal taxon with this complex repertoire of sense organs, and they exhibit evolutionary gain and loss of these sensory structures within the group. They also provide a valuable comparison to their sister clade, the Bilateria.
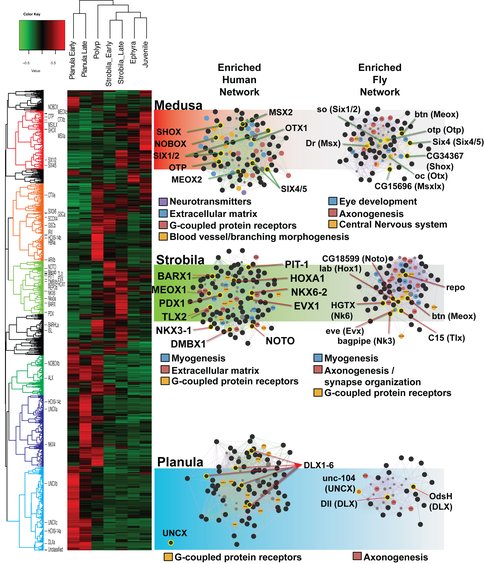
This year we published results on the combinatorial code of gene expression in sense organs using expression data from in situ hybridization and transcriptomes looking at candidate genes (Nakanishi et al. 2015). We then presented initial results of a more comprehensive analysis of transcriptomic data at the inaugural meeting of The Pan American Society for Evolutionary Developmental Biology.
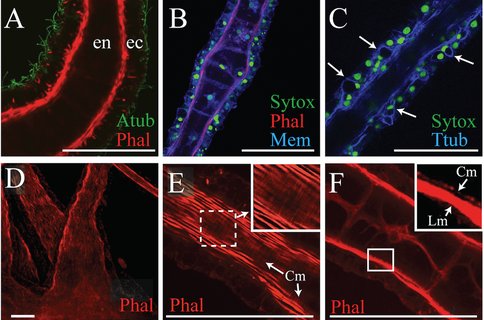
There are different tentacle types in the medusa and polyp life history stages in Aurelia. These are effectively sensory organs, but also engaged in motor response to sensation. They provide an opportunity to further study how genetic regulatory processes differentiate a suite of complicated structures. Preparatory to such studies, we characterized the differences between these tentacles of Aurelia using confocal and electron microscopy (Gold et al. 2015) and found structural evidence of a previously unrecognized motor apparatus involved in extension of the polyp tentacles, in addition to the already known mechanisms for tentacle contraction.
Publications
-
Gold, D. A., Nakanishi, N., Hensley, N. M., Cozzolino, K., Tabatabaee, M., Martin, M., … Jacobs, D. K. (2015). Structural and Developmental Disparity in the Tentacles of the Moon Jellyfish Aurelia sp.1. PLoS ONE, 10(8), e0134741. doi:10.1371/journal.pone.0134741
-
Nakanishi, N., Camara, A. C., Yuan, D. C., Gold, D. A., & Jacobs, D. K. (2015). Gene Expression Data from the Moon Jelly, Aurelia, Provide Insights into the Evolution of the Combinatorial Code Controlling Animal Sense Organ Development. PLoS ONE, 10(7), e0132544. doi:10.1371/journal.pone.0132544
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Gordon Fain
Co-Investigator
David Gold
Co-Investigator
Volker Hartenstein
Co-Investigator
Nagayasu Nakanishi
Co-Investigator
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.