2013 Annual Science Report
 University of Wisconsin
Reporting | SEP 2012 – AUG 2013
University of Wisconsin
Reporting | SEP 2012 – AUG 2013
Project 1E: Metagenomic Analysis of Novel Chemolithoautotrophic Bacterial Cultures
Project Summary
Metagenomic sequence information was obtained from two chemolithoautotrophic bacterial cultures: (1) an iron-oxidizing, nitrate-reducing culture that is capable of growth with either soluble or insoluble, mineral-bound (biotite, smectite) Fe(II) as the sole energy source; and (2) an aerobic iron/sulfur-oxidizing culture that grows with insoluble framboidal pyrite as the sole energy source. Both of these cultures carry-out novel neutral-pH lithotrophic microbial pathways, the discovery of which broadens our view of potential Fe/S based life on Earth (past and present) and other rocky planets. We hypothesize that genetic components of Fe/S oxidation identified in the metagenome of the cultures will bear resemblance to analogous components to be identified in other iron-oxidizing pure cultures being sequenced at JGI, together with existing published and unpublished information from other chemolithoautotrophic microorganisms. Identification of such genetic systems will enable comparative genomic analysis of mechanisms of extracellular phyllosilicate Fe/S redox metabolism, and facilitate development of techniques to detect the presence and expression of genes associated with chemolithotrophic Fe/S metabolism in various terrestrial environments.
Project Progress
The goal of this project is to obtain insight into the genetic/biochemical mechanisms of energy metabolism by novel chemolithoautotrophic iron/sulfur-oxidizing bacterial enrichment cultures. Identification of the functional biological components (i.e. genes and proteins) involved in these chemolithotrophic processes is critical for making a concrete linkage between biological metabolism and the generation of mineralogical and isotopic biosignatures in relation to redox gradients on Earth and other rocky planets.
Metagenomic sequence information was obtained from two chemolithoautotrophic bacterial cultures: (1) an iron-oxidizing, nitrate-reducing culture that is capable of growth with ferrous iron [Fe(II)] as the sole energy source; and (2) an aerobic iron/sulfur-oxidizing culture that grows with framboidal pyrite as the sole energy source. Both of these cultures carry-out novel neutral-pH lithotrophic microbial pathways, the discovery of which broadens our view of potential Fe/S based life on Earth (past and present) and other rocky planets.
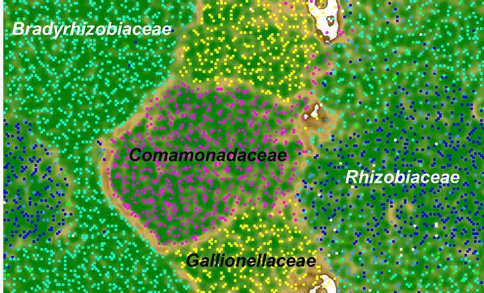
A metagenome with a total of 21.6 megabases was obtained for the Fe(II)-oxidizing, nitrate-reducing “Straub culture”, which has been show in previous studies to be capable of chemolithoautotrophic growth with soluble (Straub et al., 1996; Blöthe and Roden, 2009) or insoluble, mineral-bound (siderite, magnetite, biotite, smectite) (Weber et al., 1998; Shelobolina et al., 2012a; Xiong et al., 2013) Fe(II) as the sole energy source. Contigs longer than 3 kb were binned to different taxonomic groups (Fig. 1). Four draft reconstructed genomes were obtained from the abundant species utilizing a bioinformatics technique known as Emergent Self-Organizing Maps, abbreviated as ESOM (Dick et al., 2009; Wrighton et al., 2012; Castelle et al., 2013). Consistent with a parallel 16S rRNA gene pyrosequence library obtained for the culture, the four organisms belong to the families of Gallionellaceae, Comamonadaceae, Bradyrhizobiaceae, and Rhizobiaceae.
The draft genome of the Gallionella sp. is estimated to be >97% complete, allowing for metabolic reconstruction and comparison to its close relative Sideroxydans lithotrophicus ES-1 (Blöthe and Roden, 2009; Emerson et al., 2010). Homologs of MtoAB involved in Fe(II) oxidation by ES-1, were present in the Gallionellaceae sp. draft genome. Specifically, MtoA encodes a c-type cytochrome, and MtoB is predicted to be an outer membrane beta-barrel protein with 28 transmembrane regions, consistent with their hypothesized role in extracellular Fe(II) oxidation (Liu et al., 2012; Emerson et al., 2013). We are continuing to search for cytochromes, multicopper oxidases, and outer membrane proteins that might be involved in Fe(II) oxidation from the draft genomes of other populations in the culture. We suspect that some, if not all flanking populations, are able to oxidize Fe(II) and grow lithoautotrophically, as our data suggests that Rubisco and relevant carbon fixation genes are present in the draft genomes binned as Bradyrhizobiaceae and Rhizobiaceae. These results are consistent with new pure culture whole genome sequence information (obtained through the U.S. Department of Energy Joint Genome Institute’s Microbial Isolates sequencing program) available for three strains of Bradyrhizobium, which were recently shown to be capable of chemolithotrophic Fe(II) oxidation with either oxygen or nitrate as the electron acceptor (Shelobolina et al., 2012b; Benzine et al., 2013).
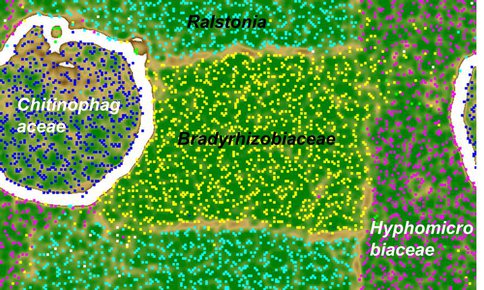
An analogous metagenomic reconstruction of organisms in a novel chemolithoautotrophic pyrite-oxidizing culture (Percak-Dennett et al., 2013) wasundertaken. In line with 16S rRNA gene library analysis, this culture is dominated by members of the Order Rhizobiales (Bradyrhizobiaceae and Hyphomicrobiaceae families) together with an organism from the genus Ralstonia and the family Chitinophagaceae (Fig. 2).
The genomes of both the Rhizobales members contain complete Rubisco pathways for autotrophic carbon fixation. Homologs of sulfur-oxidizing sox genes were recovered in all community members, with Ralstonia, Bradyrhizobiaceae, and Hyphomicrobiaceae all containing a complete sox pathway capable of thiosulfate oxidation to sulfate. A predicted outer membrane protein, homologous to the outer membrane c-type cytochrome (cyc2) of Acidithiobacillus ferrooxidans involved in Fe(II) oxidation (Yarzabal et al., 2002), was found in the Bradyrhizobiaceae draft genome. In addition, we also found cytochrome c or multicopper oxidase genes next to beta-barrel outer membrane proteins in the draft genomes of Ralstonia, Bradyrhizobiaceae, and Hyphomicrobiaceae, and therefore suspect that these genes probably play a role in extracellular electron transfer during Fe/S oxidation on the bacterial surface (Liu et al., 2012).
Further identification and characterization of the genetic systems discussed above will enable comparative genomic analysis of mechanisms of extracellular phyllosilicate Fe/S redox metabolism, and facilitate development of techniques to detect the presence and expression of genes associated with chemolithotrophic Fe/S metabolism in various terrestrial (and marine) environments.
References cited
Benzine, J., E. Shelobolina, M. Y. Xiong, D. W. Kennedy, J. P. McKinley, X. Lin, and E. E. Roden. 2013. Fe-phyllosilicate redox cycling organisms from a redox transition zone in Hanford 300 Area sediments. Front. Microbiol. 4:388.
Blöthe, M., and E. E. Roden. 2009. Composition and activity of an autotrophic Fe(II)-oxidizing, nitrate-reducing enrichment culture. Appl. Environ. Microbiol. 75:6937–6940.
Castelle, C. J., L. A. Hug, K. C. Wrighton, B. C. Thomas, K. H. Williams, D. Y. Wu et al. 2013. Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat. Commun. 4.
Dick, G. J., A. F. Andersson, B. J. Baker, S. L. Simmons, A. P. Yelton, and J. F. Banfield. 2009. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 10.
Emerson, D., J. A. Rentz, and T. Plaia. 2010. Sideroxydans lithotrophicus, gen. nov., sp. nov. and Gallionella capsiferriformans sp. nov., oxygen-dependent ferrous iron-oxidizing bacteria that grow at circumneutral pH. Int. J. Syst. Evol. Microbiol. In press.
Emerson, D., E. K. Field, O. Chertkov, K. W. Davenport, L. Goodwin, C. Munk et al. 2013. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front. Microbiol. 4:254.
Liu, J., Z. Wang, S. M. Belchik, M. J. Edwards, C. Liu, D. W. Kennedy et al. 2012. Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 3:37.
Percak-Dennett, E. M., B. J. Converse, S. He, H. Konishi, H. Xu, C. S. Chan et al. 2013. Aerobic microbial pyrite oxidation at circumneutral pH. Manuscript in preparation.
Shelobolina, E. S., H. Xu, H. Konishi, R. Kukkadapu, T. Wu, M. Blothe, and E. E. Roden. 2012a. Microbial lithotrophic oxidation of structural Fe(II) in biotite. Appl. Environ. Microbiol. 78:5746–5752.
Shelobolina, E. S., H. Konishi, H. Xu, J. Benzine, M. Xiong, T. Wu et al. 2012b. Isolation of iron-cycling organisms from illite-smectite rich rhizosphere soil. Front. Microbiol. 3:134.
Straub, K. L., M. Benz, B. Schink, and F. Widdel. 1996. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl. Environ. Microbiol. 62:1458-1460.
Weber, K. A., E. E. Roden, and F. W. Picardal. 1998. Microbially-catalyzed nitrate-dependent oxidation of solid-phase Fe(II) compounds. Environ. Sci. Technol. Submitted for publication.
Wrighton, K. C., B. C. Thomas, I. Sharon, C. S. Miller, C. J. Castelle, N. C. VerBerkmoes et al. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661-1665.
Xiong, M., E. Shelobolina, and E. Roden. 2013. Potential for microbial oxidation of ferrous iron in basaltic glass. Astrobiology Submitted for publication.
Yarzabal, A., G. Brasseur, J. Ratouchniak, K. Lund, D. Lemesle-Meunier, J. A. DeMoss, and V. Bonnefoy. 2002. The high-molecular-weight cytochrome c Cyc2 of of Acidithiobacillus ferrooxidans is an outer membrane protein. FEMS Microbiol. Lett. 209:189-195.
Publications
- Percak-Dennett, E.M., C., B.J., H., S., K., H., X., H., C., C.S., B., A., B.T. & Roden, E.E. (2013). Aerobic microbial pyrite oxidation at circumneutral pH.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Eric Roden
Project Investigator
Shaomei He
Co-Investigator
Eric Boyd
Collaborator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.3
Biochemical adaptation to extreme environments
Objective 6.2
Adaptation and evolution of life beyond Earth