2013 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2012 – AUG 2013
Massachusetts Institute of Technology
Reporting | SEP 2012 – AUG 2013
Molecular Biosignatures: Reconstructing Events by Comparative Genomics
Project Summary
Reconstructing ancient events in genome evolution provides a valuable narrative for planetary history. Phylogenetic analysis of protein families within microbial lineages can be used to detect horizontal gene transfers and the evolution of new metabolic pathways and physiologies, many of which are significant in reconstructing ancient ecologies and biogeochemical events. These gene transfers can also be used to constrain molecular clock models for early life evolution, applying principles of stratigraphy and date calibration. A better understanding of gene evolution, including partial horizontal gene transfer, is needed to improve these inferences and avoid systematic errors.
Project Progress
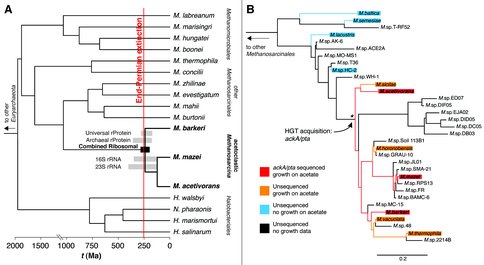
(1) In collaboration with Dan Rothman, Ed Boyle, and Roger Summons, we have further developed a geochemical model for how a horizontal gene transfer evolving a novel pathway of acetoclastic methanogenesis within methanogenic Archaea may have played a role in altering the carbon cycle of the Late Permian. We show that the sulfidic, anoxic environment of the Late Permian, in concert with increased levels of Ni in marine sediments, would favor the growth of these methanogens. We also propose that the presence of this new efficient pathway for mineralizing acetate within a fast-growing population of methanogens would have directly impacted the large organic carbon reservoir within Late Permian sediments, increasing net carbon dioxide release. Improvements to our time calibration of methanogen evolution more narrowly constrain the timing of this gene transfer event, more closely associating it with the End-Permian. We also now validate the inferred divergence times within this clock model by showing their congruence with other published time-calibrated gene transfers from Archaea to Cyanobacteria and eukaryotes.
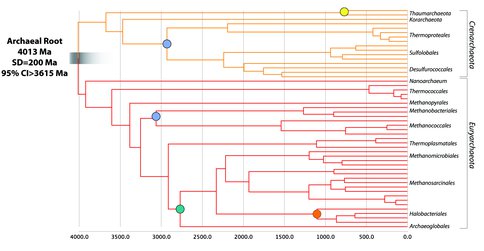
(2) Elaborating this principle, we have also developed a theory of “gene transfer stratigraphy,” which can use a set of gene transfers to impose relative time calibrations within phylogenetic trees. We have identified numerous horizontal transfers within archaeal, bacterial, and eukaryotic genomes that have utility for this novel approach. Preliminary results using this new methodology for the time-calibration of Archaea support models for an early origin of methanogenesis (>3.0 Ga), and a very early origin of Archaea, most likely ~4.0 Ga, and certainly earlier than 3.6 Ga. These models support a timing of the Last Common Ancestor and Origin of Life very early in planetary history, before the time of the proposed “Late Heavy Bombardment”, and provide evidence against models of impact frustration for the Origin of Life on the early Earth. We anticipate that improvements in molecular clock models implementing relative gene transfer constraints will permit more valuable calibrations to refine these models and further improve date estimates.
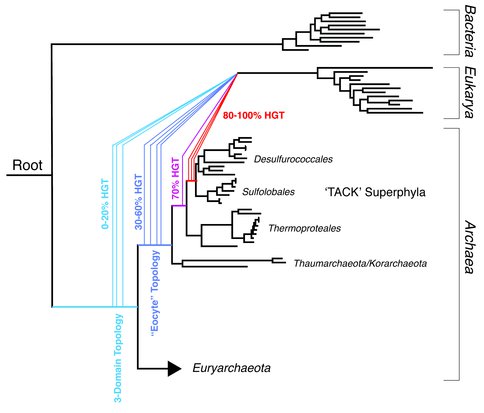
(3) In an attempt to improve phylogenetic analyses of gene trees, we have also investigated the presence of partial horizontal gene transfer within sequence datasets, and their impact on the inference of ancient evolutionary events. Using simulations, we show that if a transferred gene recombines with a vertically inherited copy of the gene, a phylogenetic reconstruction of the gene history will place the recipient lineage incorrectly. We identify partial gene transfer events within several important genes in the eukaryotic lineage, and propose that these undetected transfers are the cause of an artifact that seemingly places eukaryotes as a derived archaeal lineage within some analyses. We further show that these partial gene transfers are extensive within a diverse set of archaeal lineages, and may be a major source of uncertainty and error in phyogenomic reconstructions.
Publications
-
Rothman, D. H., Fournier, G. P., French, K. L., Alm, E. J., Boyle, E. A., Cao, C., & Summons, R. E. (2014). Methanogenic burst in the end-Permian carbon cycle. Proceedings of the National Academy of Sciences, 111(15), 5462–5467. doi:10.1073/pnas.1318106111
- Andam, C., Fournier, G.P. & Gogarten, J.P. Ancient horizontal gene transfer and the last commonancestors. In: Koonin, E., Mulkidjanian, A. & Lankenau, D. (Eds.). LUCA: The Last Universal Cellular Ancestor of life. New York: Springer Verlag.
- Fournier, G.P., Freese, P. & Alm, E.J. (2013). Re-evaluating the Eocyte Hypothesis in Light of Intragenic Horizontal Gene Transfer. PLOS Biology.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Eric Alm
Co-Investigator
Edward Boyle
Co-Investigator
Greg Fournier
Co-Investigator
Katherine French
Co-Investigator
Dan Rothman
Co-Investigator
Roger Summons
Co-Investigator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.
Objective 4.3
Effects of extraterrestrial events upon the biosphere
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 6.1
Effects of environmental changes on microbial ecosystems