2013 Annual Science Report
 Massachusetts Institute of Technology
Reporting | SEP 2012 – AUG 2013
Massachusetts Institute of Technology
Reporting | SEP 2012 – AUG 2013
Early Animals: Sensory Systems and Combinatorial Codes
Project Summary
Understanding the evolution of integrated sensory organs, such as the eyes, ears and nose that develop in concert on our heads, is fundamental to understanding animal complexity, as these are the features that permit movement and the environmental responses that characterize animals. We are looking at understudied early branches of the animal family tree—including the jellyfish Aurelia and the annelid worm Neanthes—to understand how the genetic regulation of sensory organs is conserved in some cases and evolves in others. Comparisons of developmental regulation in different clades reveal how similar gene networks can be differentially modified and deployed, permitting the evolution of complex sensory systems. The application of genomic methods greatly enhances our ability to pursue these questions.
Project Progress
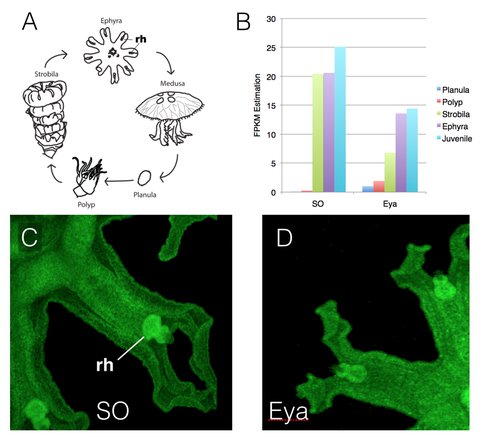
One perspective on the genetic control of sense organ development is that they are regulated by a combinatorial code, where expression of some genes is shared across multiple sensory structures, while the expression of other genes is specific to particular organs. Jellyfish are an excellent model for this work in that they are more complex in life history than the bilaterally symmetric organisms (Bilateria). Additionally, sense organs may have been gained and lost, and are deployed in different combinations, across the Cnidaria (the larger group to which the scyphozoan jellyfish belong). Thus, comparisons between jellyfish and bilaterian sense organ development should help elucidate the combinatorial code controlling sense organ development and thereby reveal controls on the evolution of complex organ sytems in multicellular animals. Viewed in this light, tinkering with the terminal features of the gene network could permit the conversion of one sensory modality to another in evolution. By contrast, regulatory networks are often presented as canonical and invariant, especially when studied exclusively in the Bilateria to which we, and most model systems, belong. This is the case with eye development, where the developmental genetic regulatory structure has often been presented as evolutionarily invariant. This is not the case when these genes are explored in the jellyfish. In Aurelia we find that, although many of the same genes that function in bilaterian eye development are present, not all are, and those present are not all expressed in the same stereotypic way as they are across Bilateria. Some of the genes that function in the development of multiple sensory structures appear conserved in function. On the other hand, regulatory components long thought to be critical and canonical to bilaterian eye development are absent from, or not expressed in, cnidarian sensory structures. These include the PAX genes, long thought to be critical “master regulators” of bilaterian eye specification. This work, now in review at BMC Evolutionary Biology, strongly suggests that the evolution of the combinatorial code for sense organ development has been critical to the potential for modifying sensory structures in the Cnidaria. Thus this combinatorial code model may provide an understanding of how complex structures evolved and how developmental regulation was assembled.
Many sensory structures develop from—or are associated with—appendages, and not surprisingly, there are some components of development regulation that are common to sensory structures and appendages. We published work with similar reference to combinatorial codes involving appendage evolution in the polychaete worm Neanthes (Winchell et al. 2013). This work examines the expression of LIM regulatory genes in the development of the distinctive appendages of polychaete worms. This effort illuminates the differential expression of conserved regulatory elements in polychaete appendages relative to the more typically studied arthropod appendages. Related work compares bilaterian and cnidarian stem cell function (Gold et al. 2013; Takashima et al. 2013). Stem cells appear not to function in the same manner in cnidarians as they do in bilaterians. Thus, stem cell function, although thought to be fundamentally important, may not be ancestrally conserved to the degree some might anticipate.
To further these lines of research with our collaborators, we are developing genomic resources for Aurelia. Alongside the genome, we have sequenced the transcriptome at all major developmental stages for Aurelia using RNA-Seq. This has immediately revealed dramatic differentiation of sensory genes through the life cycle. We are pursuing a wide range of analyses, and we anticipate submission of additional publications within the coming months as we continue to analyze this new wealth of data.
Publications
-
Gold, D. A., & Jacobs, D. K. (2012). Stem cell dynamics in Cnidaria: are there unifying principles?. Dev Genes Evol, 223(1-2), 53–66. doi:10.1007/s00427-012-0429-1
-
Hartenstein, V., & Jacobs, D. (2013). Developmental Plasticity, Straight from the Worm’s Mouth. Cell, 155(4), 742–743. doi:10.1016/j.cell.2013.10.038
-
Takashima, S., Gold, D., & Hartenstein, V. (2012). Stem cells and lineages of the intestine: a developmental and evolutionary perspective. Dev Genes Evol, 223(1-2), 85–102. doi:10.1007/s00427-012-0422-8
-
Winchell, C. J., & Jacobs, D. K. (2013). Expression of the Lhx genes apterous and lim1 in an errant polychaete: implications for bilaterian appendage evolution, neural development, and muscle diversification. EvoDevo, 4(1), 4. doi:10.1186/2041-9139-4-4
- Nakanishi, N., Camara, A., Yuan, D., Gold, D.A. & Jacobs, D.K. (Submitted). Sine Oculus & Eyes Absent expression is conservative in the cnidarian Aurelia while Pax genes appear evolutionarily labile, consistent with the evolution of combinatorial code that specifies animal sense organs. BMC Biology.
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Gordon Fain
Co-Investigator
David Gold
Co-Investigator
Volker Hartenstein
Co-Investigator
David Jacobs
Co-Investigator
Nagayasu Nakanishi
Co-Investigator
Chris Winchell
Co-Investigator
-
RELATED OBJECTIVES:
Objective 4.1
Earth's early biosphere.
Objective 4.2
Production of complex life.