2013 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2012 – AUG 2013
Georgia Institute of Technology
Reporting | SEP 2012 – AUG 2013
Experimental Evolution and Genomic Analysis of an E. Coli Containing a Resurrected Ancestral Gene
Project Summary
In order to study the historical pathways and modern mechanisms of protein evolution in a complex cellular environment, we combined ancestral sequence reconstruction with experimental evolution. Our first goal was to identify how ancestral states of a protein effect cellular behavior by directly engineering an ancient gene inside a modern genome. We could then identify the evolutionary steps of this organism harboring the ancient gene by subjecting it to laboratory evolution, and directly monitoring the resulting changes within the integrated ancient gene as well as the rest of the host genome.
Project Progress
Our ability to understand how the environment may change a microbial ecosystem or how life adapts and evolves to environmental challenges requires that we observe evolution in action. This is difficult, however, because evolution occurs across long time-scales. Through a synthesis of tools drawn from phylogenetics, molecular evolution and synthetic biology, we can now access and study the ancestral states of modern genes and proteins. This combinatorial approach is named “ancestral sequence resurrection”. In its simplest terms, this approach allows us to infer ancestral gene and protein sequences through phylogeny, followed by synthesis of these sequences in the laboratory and empirically testing hypotheses about how proteins evolved into their specific function (Thornton 2004).
In order to study the historical pathways and modern mechanisms of protein evolution in a complex cellular environment, we combined ancestral sequence reconstruction with experimental evolution. Our first goal was to identify how ancestral states of a protein effect cellular behavior by directly engineering an ancient gene inside a modern genome. We could then identify the evolutionary steps of this organism harboring the ancient gene by subjecting it to laboratory evolution, and directly monitoring any resulting changes within the integrated ancient gene and the rest of the host genome.
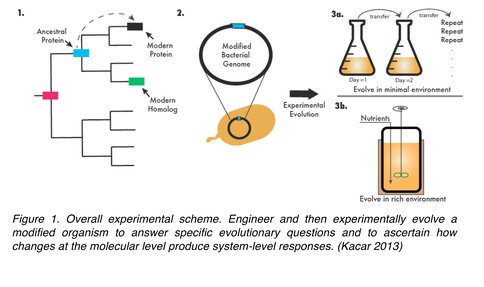
This research was carried out in four steps (Figure 1):
1. Resurrection of an ancient gene in a modern bacterial genome (previously completed)
2. Experimental evolution of bacteria harboring the ancient gene (previously completed)
3. High throughput gene and genomic sequencing of the evolved bacterium (completed during this progress year)
4. Biochemical analysis of the evolved protein and protein interaction networks (On-going)
Resurrection of an ancient gene in a bacterial genome
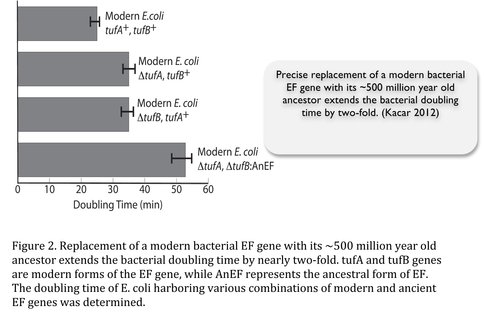
We opted to focus on a previously resurrected ancestral protein, Elongation Factor (EF) (Gaucher 2008). EF is an essential and a universal protein; carries a GTP, and primarily functions to deliver aminoacylated tRNAs to the ribosome. In addition, EF serves as a hub protein, interacting with ~100 partners, including chaperons, metabolic enzymes, structural proteins and others (Kacar 2012, 2013). Addressing our first goal. We have engineered an E. coli by replacing its endogenous EF gene with a ~500 million year old EF gene. The replacement caused the bacteria to be maladapted (observed by a two-fold increase in bacterial doubling time) (Figure 2).
Utilizing this struggle as a driver for evolution, we evolved the sick bacteria for thousands of generations in the laboratory, and monitored the adaptive pathways of the organism hosting the ancient gene through genomics and fitness measurements. We sought the answers for the following questions in particular: Will the ancient protein evolve exactly into its modern descendant, or in what other ways the bacteria and the ancient gene will co-adapt? The results shed light on our understanding of how life evolves and the deterministic and random forces guiding evolution, in particular, allows us to demonstrate the diversity of molecular level evolutionary innovations that lead to organismal survival.
Experimental evolution and whole genome sequencing
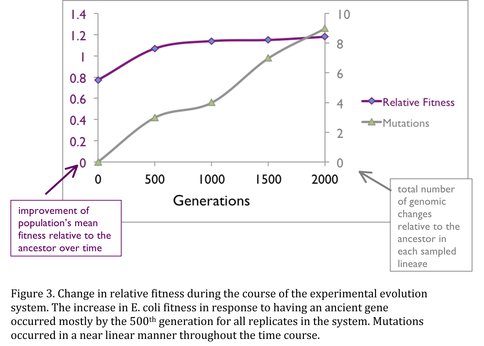
Next, the E. coli strain harboring an ancient EF was subjected to long-term laboratory evolution in minimal growth media via serial transfers in replicate lines (Elena 2003). To understand how bacteria responded to the environment and selective pressures, we monitored the changes in bacterial phenotype and genotype through fitness experiments, as well as next-generation genome sequencing (Figure 3). Various genomic and phenotypic analyses demonstrate that organismal evolution follows two hierarchically distinct levels of adaptation. In one level, bacterial lineages up-regulated the gene expression level of the ancient EF. At the other level, evolutionary constraints acted on protein-protein interaction networks: Although there is no change in the protein-coding region or the expression levels of the ancient EF for one lineage, the EF protein-protein interaction network accumulated mutations. Specifically, the proteins interacting with EF accumulated loss-of-function mutations, which in return were beneficial for the organism. Results demonstrate that adaptation via regulation of gene expression and protein-protein interaction networks individually contribute to organismal evolution. The results of this work are currently being written for publication, and below we outline the on-going biochemical work that we are using to dissect the molecular level changes in the evolved EF protein-protein interaction network.
Genomic and biochemical analysis of the evolved protein-protein interaction network
Null alleles (in genes infB and nusA) are detected in one evolved lineage for the EF protein-protein interaction network. These mutations result in a loss of function in Initiation Factor (IF2) protein (an essential protein involved in the ribosomal machinery) and a nusA protein (a transcription regulatory protein), and are known to interact with EF (Sacerdot 1984, Bylund 2001, Arifuzzaman 2006, Kacar 2013). So far, our data shows that the loss of function in nusA increases bacterial fitness. Currently we are investigating how this mutation affects the biochemical interaction between nusA and wild-type EF and ancient EF. Further, through our collaboration with the RiboCore Center (Uppsala University, Sweden), the changes of the mutant IF2 are and will be investigated structurally, functionally and computationally. Further, various results obtained from this work are setting the foundations for experimental systems being developed to explore the evolutionary forces that drive protein function, how proteins interact with each other, and the functional, structural and historical constraints that shape biological evolution.
References
Arifuzzaman, M., M. Maeda, A. Itoh, et al. (2006). “Large-scale identification of protein-protein interaction of Escherichia coli K-12.” Genome Research 16(5): 686-691.
Bylund, G. O., J. M. Lovgren and P. M. Wikstrom (2001). “Characterization of mutations in the metY-nusA-infB operon that suppress the slow growth of a DeltarimM mutant.” Journal of Bacteriology 183(20): 6095-6106.
Dean, A. M. and J. W. Thornton (2007). “Mechanistic approaches to the study of evolution: the functional synthesis.” Nature Reviews Genetics 8(9): 675-688.
Elena, S. F. and R. E. Lenski (2003). “Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation.” Nature Reviews Genetics 4(6): 457-469.
Gaucher, E. A., S. Govindarajan and O. K. Ganesh (2008). “Palaeotemperature trend for Precambrian life inferred from resurrected proteins.” Nature 451(7179): 704-707.
Kacar, B. and E. A. Gaucher (2013). “Experimental evolution of protein-protein interaction networks.” Biochemical Journal 453(3): 311-319.
Kacar, B., Gaucher, EA. (2012) “Towards the Recapitulation of Ancient History in the Laboratory: Combining Synthetic Biology with Experimental Evolution.” Artificial Life, 13:11-18.
Sacerdot, C., P. Dessen, J. W. Hershey, J. A. Plumbridge and M. Grunberg-Manago (1984). “Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors.” Proc Natl Acad Sci U S A 81(24): 7787-7791.
Publications
-
Cacan, E., Kratzer, J. T., Cole, M. F., & Gaucher, E. A. (2013). Interchanging Functionality Among Homologous Elongation Factors Using Signatures of Heterotachy. Journal of Molecular Evolution, 76(1-2), 4–12. doi:10.1007/s00239-013-9540-9
-
Ingles-Prieto, A., Ibarra-Molero, B., Delgado-Delgado, A., Perez-Jimenez, R., Fernandez, J. M., Gaucher, E. A., … Gavira, J. A. (2013). Conservation of Protein Structure over Four Billion Years. Structure, 21(9), 1690–1697. doi:10.1016/j.str.2013.06.020
-
Kaçar, B., & Gaucher, E. A. (2013). Experimental evolution of protein–protein interaction networks. Biochemical Journal, 453(3), 311–319. doi:10.1042/bj20130205
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Betül Kacar
Collaborator
Joshua Stern
Collaborator
-
RELATED OBJECTIVES:
Objective 3.4
Origins of cellularity and protobiological systems
Objective 4.1
Earth's early biosphere.
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 6.1
Effects of environmental changes on microbial ecosystems
Objective 6.2
Adaptation and evolution of life beyond Earth