2013 Annual Science Report
 Georgia Institute of Technology
Reporting | SEP 2012 – AUG 2013
Georgia Institute of Technology
Reporting | SEP 2012 – AUG 2013
An Atomic Level Description of the Specific Interactions Between Nascent Peptide and Ribosome Exit Tunnel
Project Summary
The ribosome exit tunnel is an ancient path that must be traveled by all peptides/proteins synthesized by the ribosome. We have synthesized peptolides and demonstrated their potential as probes to decipher the interaction between the nascent peptide and the exit tunnel. This study has furnished vital information about the path of travel of peptides attached to the flag-pole moiety of a ketolide. In continuation of our study, we have designed and synthesized a second generation of hybrid molecules taking inspiration from peptide sequences that are known to naturally stall translation. We will characterize the interactions of these stall peptolides with the ribosome exit tunnel using the tools we have developed during our investigation of the peptolides.
Project Progress
Abstract
The ribosome exit tunnel is an ancient path that must be traveled by all peptides/proteins synthesized by the ribosome. We have synthesized peptolides and demonstrated their potential as probes to decipher the interaction between the nascent peptide and the exit tunnel. This study has furnished vital information about the path of travel of peptides attached to the flag-pole moiety of a ketolide. In continuation of our study, we have designed and synthesized a second generation of hybrid molecules taking inspiration from peptide sequences that are known to naturally stall translation. We will characterize the interactions of these stall peptolides with the ribosome exit tunnel using the tools we have developed during our investigation of the peptolides.
Narrative
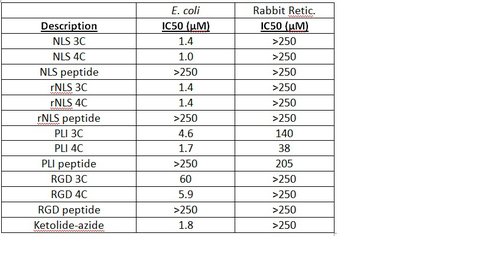
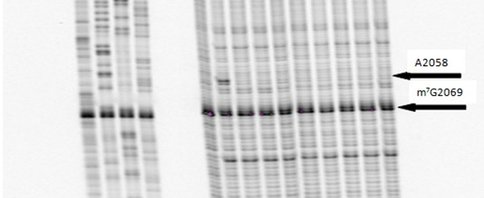
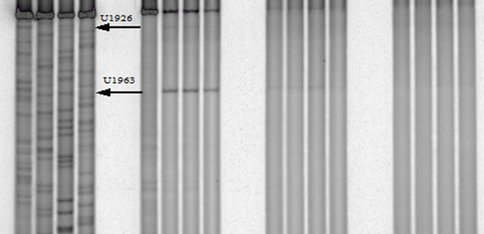
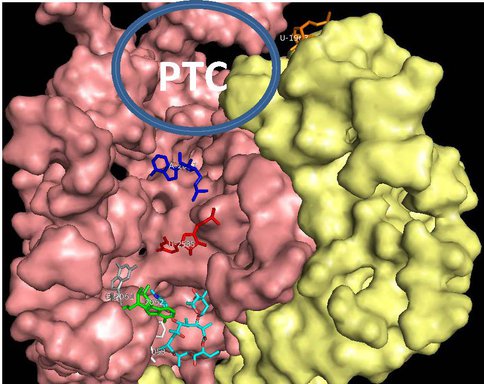
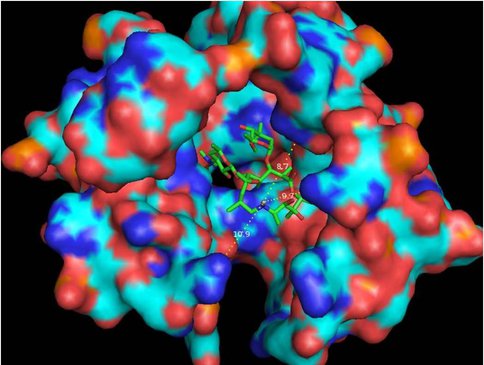
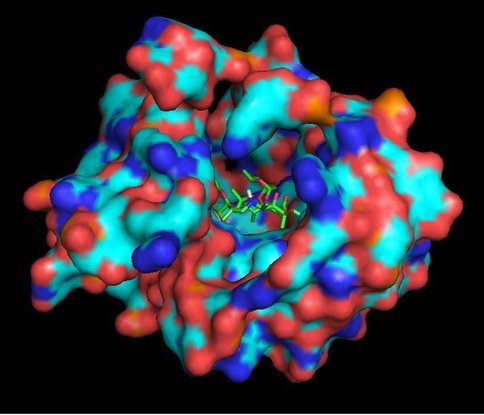
One of our primary interests is to elucidate the nature of the interaction between the ribosome peptide exit tunnel and the nascent peptide before breeching the extraribosomal interface. In doing so, we will be able to gain a better understanding of the molecular basis for the choice of nucleic acid as exit tunnel construction materials. Using molecular probes – peptolides – that we have developed, we have characterized the interaction between varied peptide sequences and key residues within the exit tunnel and the space adjoining the entrance to the exit tunnel and the peptidyl transferase site. This was accomplished by cell-free translation inhibition assays (Table 1), footprinting experiments of the ribosomal RNA (rRNA) through dimethyl sulfate (DMS) and 1-cyclohexyl-(2-morpholino-ethyl)carbodiimide metho-p-toluene sulfonate (CMCT) modification in the presence of the peptolides (Figs. 1 & 2). Our footprinting data strongly suggest that the peptide moieties of all the peptolide studied preferred a path of travel where they are oriented back to the peptidyl transferase site. This observation is consistent with crystallographic data on telithromycin (ketek), the ketolide template for all our peptolides, showing that it could adopt a conformation which has its flag-pole moiety oriented toward the peptidyl transferase site.1-3 This path, shown in Figure 3, covers the portion of the exit tunnel from the PTC through to the macrolide binding site, approximately a quarter of the full length of the tunnel.
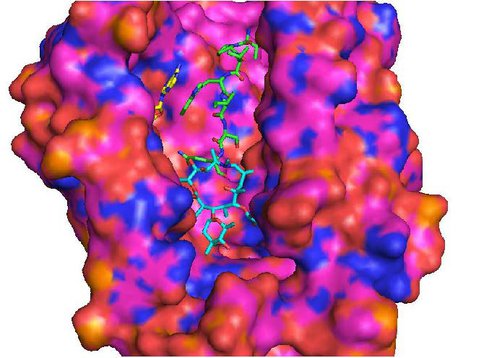
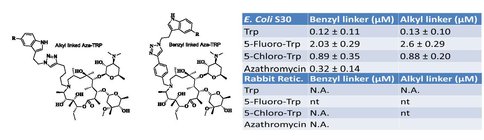
To cover the remaining part of the exit tunnel, we have designed another generation of peptolides built from an azalide azathromycin, using its endocyclic amine group (N10) as a connection point to the peptide moiety. By replacing the telithromycin scaffold with that of azithromycin, the flexibility of the flag-pole moiety is removed, thus selectively presenting the attached peptide further down the tunnel. Crystal structures of both H. marismortui and T. thermophilus with azithromycin bound show that the proposed site of peptide attachment, N10, has an expansive space that would permit modification (Figs. 4, 5). The peptide probe in this series will be modeled after SecM, a naturally occurring peptide that stalls translation.4 As shown in Fig. 6, W155 seems to interact with A752 of the 23S in a pi-pi stack. We have synthesized series of model indole-azalide compounds to probe the SecM W155-23S A752 stacking interaction. The structures of the model compounds, as well as preliminary translation inhibition data, can be seen in Table 2. Our preliminary translation data support the importance of the SecM W155-23S A752 stacking as unsubstituted indole compound has improved the IC50 (ca. 3-fold) relative to the starting azathromycin. These compounds are now being investigated in footprinting assay to conclusively demonstrate the SecM W155-23S A752 stacking interaction. Once confirmed, the best model compound will be the template for the synthesis of the 2nd generation peptolides.
References
1) Berisio, R.; Harms, J.; Schluenzen, F.; Zarivach, R.; Hansen, H. A.; Fucini, P.; Yonath, A. J. Bacteriol. 2003, 185, 4276-4279.
2) Tu, D.; Blaha, G.; Moore, P. B.; Steitz, T. A. Cell 2005, 121, 257–270.
3) Dunkle, J. A.; Xiong, L.; Mankin, A. S.; Cate, J. H. D. PNAS, 2010, 107, 17152-17157.
4) Gumbert, J.; Schreiner, G.J.; Wilson, D.N.; Beckmann, R.; and Schulten, K. Biophys. J. 2012, 103, 331−341
Presentations
1) Canzoneri, J.C., Washington, A.Z., Benicewicz, D.B., Oyelere, A.K.; Exploring the Ribosome with Peptolides, presented at the 2011 Southeast Regional Meeting of the ACS (SERMACS),October 27, 2011, Richmond, VA. Oral Presentation
2) Canzoneri, J.C., Washington, A.Z., Benicewicz, D.B., Oyelere, A.K.; Exploring the Ribosome with Peptolides, presented at the 2012 Astrobiology Science Conference (AbSciCon), April 18, 2012, Atlanta, GA. Oral Presentation
3) Washington, A.Z., Canzoneri, J.C., Benicewicz, D.B., Oyelere, A.K.; Synthesis of Peptolides for Use as Molecular Probes, presented at the 2012 Astrobiology Science Conference (AbSciCon), April 18, 2012, Atlanta, GA. Poster Presentation
4) Washington, A.Z., Tapadar, S., Oyelere, A.K.; Interrogating potential interactions fo macrolide-stall peptide hybrids and the ribosomal exit tunnel, presented at the 2013 Southeast Regional Meeting of the ACS (SERMACS), November 13, 2013, Atlanta, GA. Poster Presentation.
Publications
-
Hsiao, C., Lenz, T. K., Peters, J. K., Fang, P-Y., Schneider, D. M., Anderson, E. J., … Dean Williams, L. (2013). Molecular paleontology: a biochemical model of the ancestral ribosome. Nucleic Acids Research, 41(5), 3373–3385. doi:10.1093/nar/gkt023
-
Petrov, A. S., Bernier, C. R., Gulen, B., Waterbury, C. C., Hershkovits, E., Hsiao, C., … Williams, L. D. (2014). Secondary Structures of rRNAs from All Three Domains of Life. PLoS ONE, 9(2), e88222. doi:10.1371/journal.pone.0088222
-
Petrov, A. S., Bernier, C. R., Hershkovits, E., Xue, Y., Waterbury, C. C., Hsiao, C., … Williams, L. D. (2013). Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Research, 41(15), 7522–7535. doi:10.1093/nar/gkt513
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Subhasish Tapadar
Collaborator
Arren Washington
Collaborator
-
RELATED OBJECTIVES:
Objective 3.2
Origins and evolution of functional biomolecules