2013 Annual Science Report
 Arizona State University
Reporting | SEP 2012 – AUG 2013
Arizona State University
Reporting | SEP 2012 – AUG 2013
Stoichiometry of Life, Task 3b: Ancient Records - Genomic
Project Summary
Task 3b team members are involved in deciphering genomic records of modern organisms as a way to understand how life on Earth evolved. At its core, this couples the integrated measurement and modeling of evolutionary mechanisms that drove the differences between extant genomes (and metagenomes), with experimental data on how environmental dynamics might have shaped these differences across geological timescales. This goal draws from team members’ expertise encompassing theoretical and computational biology, microbial evolution, and studying life in both extreme and dynamic environments across the planet.
Project Progress
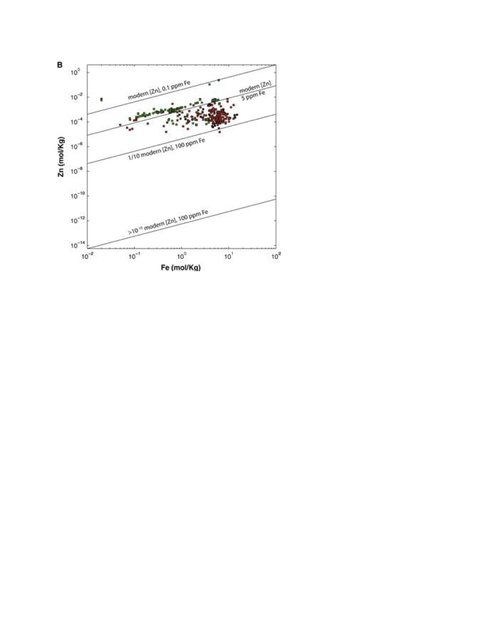
Team members for Task 3b have been involved in several noteworthy accomplishments. DuPont was a co-author on two recent publications, one in Nature Geoscience, on the role of zinc in the early biosphere (this publication bridges with Task 3a). Eukaryotic metallomes in particular may have been particularly dependent on zinc, and its bio-availability may have constrained (or driven) the evolution of early eukaryotes. Team members Boyd and Raymond continue to make advances in studying the genomic record of how life adapts to environmental extremes, with Yellowstone National Park as a focus for field studies. Several manuscripts are now in preparation, and this work was the foundation for a paper by Raymond on using signatures derived from metagenomes to better understand microbial evolution. These projects substantially advanced the doctoral progress of ASU students Alsop and Kellom. Though the project will draw to a close in the near future, collaborations between team members will continue to drive new ways to integrate their interests and expertise into a stronger understanding of how the availability of key elements drove biological innovation and evolution.
Publications
-
Alsop, E. B., & Raymond, J. (2013). Resolving Prokaryotic Taxonomy without rRNA: Longer Oligonucleotide Word Lengths Improve Genome and Metagenome Taxonomic Classification. PLoS ONE, 8(7), e67337. doi:10.1371/journal.pone.0067337
-
Robbins, L. J., Lalonde, S. V., Saito, M. A., Planavsky, N. J., Mloszewska, A. M., Pecoits, E., … Konhauser, K. O. (2013). Authigenic iron oxide proxies for marine zinc over geological time and implications for eukaryotic metallome evolution. Geobiology, 11(4), 295–306. doi:10.1111/gbi.12036
-
Scott, C., Planavsky, N. J., Dupont, C. L., Kendall, B., Gill, B. C., Robbins, L. J., … Lyons, T. W. (2012). Bioavailability of zinc in marine systems through time. Nature Geosci, 6(2), 125–128. doi:10.1038/ngeo1679
-
PROJECT INVESTIGATORS:
-
PROJECT MEMBERS:
Christopher Dupont
Co-Investigator
Janet Siefert
Co-Investigator
Eric Alsop
Collaborator
Eric Boyd
Collaborator
Matthew Kellom
Collaborator
-
RELATED OBJECTIVES:
Objective 5.1
Environment-dependent, molecular evolution in microorganisms
Objective 5.2
Co-evolution of microbial communities
Objective 5.3
Biochemical adaptation to extreme environments